1QIB
 
 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
1QIC
 
 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
1QIA
 
 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
1OWT
 
 | | Structure of the Alzheimer's disease amyloid precursor protein copper binding domain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Barnham, K.J, McKinstry, W.J, Multhaup, G, Galatis, D, Morton, C.J, Curtain, C.C, Williamson, N.A, White, A.R, Hinds, M.G, Norton, R.S, Beyreuther, K, Masters, C.L, Parker, M.W, Cappai, R. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alzheimer's Disease Amyloid Precursor Protein Copper Binding Domain. A REGULATOR OF NEURONAL COPPER HOMEOSTASIS.
J.Biol.Chem., 278, 2003
|
|
3TGK
 
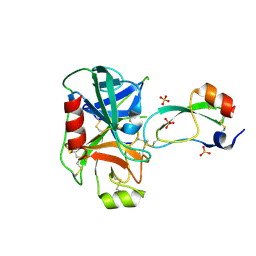 | | TRYPSINOGEN MUTANT D194N AND DELETION OF ILE 16-VAL 17 COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Medina, N, Cahoon, M, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-19 | | Release date: | 2001-07-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
1EEP
 
 | | 2.4 A RESOLUTION CRYSTAL STRUCTURE OF BORRELIA BURGDORFERI INOSINE 5'-MONPHOSPHATE DEHYDROGENASE IN COMPLEX WITH A SULFATE ION | | Descriptor: | INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | McMillan, F.M, Cahoon, M, White, A, Hedstrom, L, Petsko, G.A, Ringe, D. | | Deposit date: | 2000-02-01 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of Borrelia burgdorferi inosine 5'-monophosphate dehydrogenase: evidence of a substrate-induced hinged-lid motion by loop 6.
Biochemistry, 39, 2000
|
|
5KPM
 
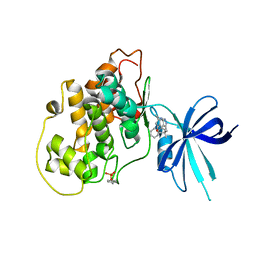 | | Glycogen Synthase Kinase 3 beta Complexed with BRD3731 | | Descriptor: | (4~{S})-3-(2,2-dimethylpropyl)-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPL
 
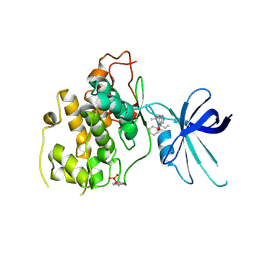 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0705 | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPK
 
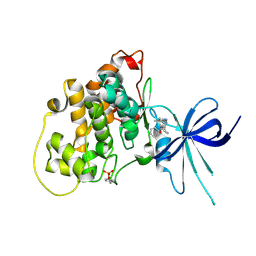 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0209 | | Descriptor: | (4~{S})-3-cyclopropyl-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
1F5R
 
 | | RAT TRYPSINOGEN MUTANT COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, TRYPSIN II, ... | | Authors: | Pasternak, A, White, A, Cahoon, M, Ringe, D, Hedstrom, L. | | Deposit date: | 2000-06-15 | | Release date: | 2001-07-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
1F7Z
 
 | | RAT TRYPSINOGEN K15A COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Medina, N, Cahoon, M, Ringe, D, Hedstrom, L. | | Deposit date: | 2000-06-28 | | Release date: | 2001-07-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
1FY8
 
 | | CRYSTAL STRUCTURE OF THE DELTAILE16VAL17 RAT ANIONIC TRYPSINOGEN-BPTI COMPLEX | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Ringe, D, Hedstrom, L. | | Deposit date: | 2000-09-28 | | Release date: | 2000-11-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
1FDJ
 
 | | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE FROM RABBIT LIVER | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE, ... | | Authors: | Blom, N.S, White, A, Sygusch, J. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction intermediates of Rabbit liver D-fructose 1,6-bisphosphate Aldolase
To be Published
|
|
1F5T
 
 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT) COMPLEXED WITH NICKEL AND DTXR CONSENSUS BINDING SEQUENCE | | Descriptor: | 43MER DNA CONTAINING DXTR CONSENSUS BINDING SEQUENCE, DIPHTHERIA TOXIN REPRESSOR, NICKEL (II) ION | | Authors: | Chen, S, White, A, Love, J, Murphy, J.R, Ringe, D. | | Deposit date: | 2000-06-15 | | Release date: | 2000-09-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Methyl groups of thymine bases are important for nucleic acid recognition by DtxR.
Biochemistry, 39, 2000
|
|
2YDK
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 2-(CARBAMOYLAMINO)-5-PHENYL-N-[(3S)-PIPERIDIN-3-YL]THIOPHENE-3-CARBOXAMIDE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
2YDI
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 5-[4-(2-DIMETHYLAMINOETHYLOXY)PHENYL]-2-UREIDO-THIOPHENE-3-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-21 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
2YDJ
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 5-(3-fluorophenyl)-N-[(3S)-3-piperidyl]-3-ureido-thiophene-2-carboxamide, PHOSPHATE ION, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-01-25 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
