3TEK
 
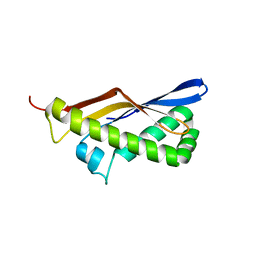 | | ThermoDBP: a non-canonical single-stranded DNA binding protein with a novel structure and mechanism | | Descriptor: | ThermoDBP-single stranded DNA binding protein | | Authors: | White, M.F, Paytubi, S, Liu, H, Graham, S, McMahon, S.A, Naismith, J.H. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7QQK
 
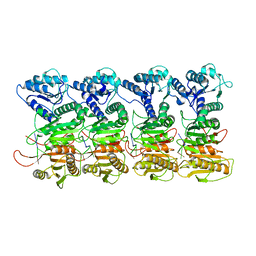 | | TIR-SAVED effector bound to cA3 | | Descriptor: | RNA (5'-R(P*AP*AP*A)-3'), TIR_SAVED fusion protein | | Authors: | Spagnolo, L, White, M.F, Hogrel, G, Guild, A. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cyclic nucleotide-induced helical structure activates a TIR immune effector.
Nature, 608, 2022
|
|
8QJK
 
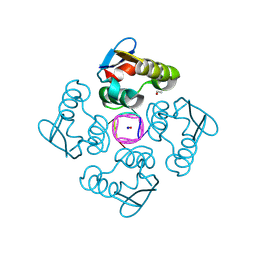 | | Structure of the cytoplasmic domain of csx23 from Vibrio cholera in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | ACETYL GROUP, Cyclic tetraadenosine monophosphate (cA4), SODIUM ION, ... | | Authors: | McMahon, S.A, McQuarrie, S, Gloster, T.M, Gruschow, S, White, M.F. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | CRISPR antiphage defence mediated by the cyclic nucleotide-binding membrane protein Csx23.
Nucleic Acids Res., 52, 2024
|
|
2X5F
 
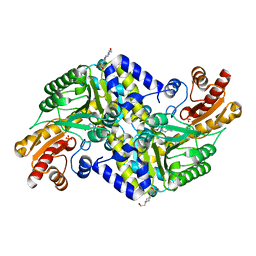 | | Crystal structure of the methicillin-resistant Staphylococcus aureus Sar2028, an aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate dependent aminotransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASPARTATE_TYROSINE_PHENYLALANINE PYRIDOXAL-5' PHOSPHATE-DEPENDENT AMINOTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3F
 
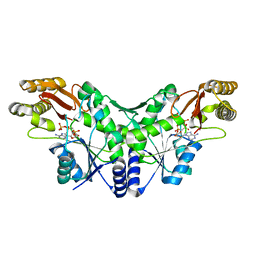 | | Crystal Structure of the Methicillin-Resistant Staphylococcus aureus Sar2676, a Pantothenate Synthetase. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PANTHOTHENATE SYNTHETASE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
6YUD
 
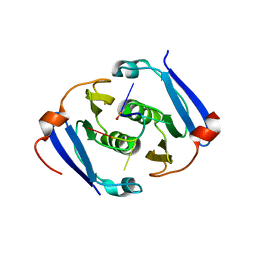 | | Structure of Csx3/Crn3 from Archaeoglobus fulgidus in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), Uncharacterized protein AF_1864 | | Authors: | McQuarrie, S, Gloster, T.M, White, M.F, Graham, S, Athukoralage, J.S, Gruschow, S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.
Elife, 9, 2020
|
|
8ANE
 
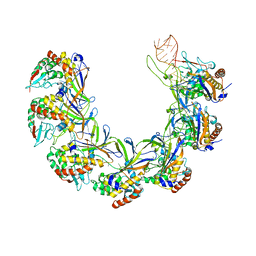 | | Structure of the type I-G CRISPR effector | | Descriptor: | Cas7, RNA (66-MER) | | Authors: | Shangguan, Q, Graham, S, Sundaramoorthy, R, White, M.F. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the type I-G CRISPR effector.
Nucleic Acids Res., 50, 2022
|
|
8B2X
 
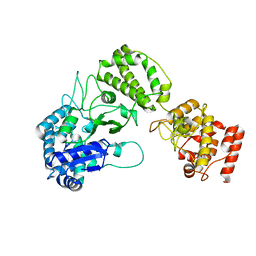 | |
2X48
 
 | | ORF 55 from Sulfolobus islandicus rudivirus 1 | | Descriptor: | CAG38821, PHOSPHATE ION | | Authors: | Oke, M, Carter, L, Johnson, K.A, Liu, H, Mcmahon, S, Naismith, J.H, White, M.F. | | Deposit date: | 2010-01-28 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X0O
 
 | | Apo structure of the Alcaligin biosynthesis protein C (AlcC) from Bordetella bronchiseptica | | Descriptor: | ALCALIGIN BIOSYNTHESIS PROTEIN, SULFATE ION | | Authors: | Johnson, K.A, Schmelz, S, Kadi, N, Mcmahon, S.A, Oke, M, Liu, H, Carter, L.G, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2009-12-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3E
 
 | | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III, FabH from Pseudomonas aeruginosa PAO1 | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 3 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5Q
 
 | | Crystal Structure of Hypothetical protein sso1986 from Sulfolobus solfataricus P2 | | Descriptor: | SSO1986 | | Authors: | Oke, M, Carter, L, Johnson, K.A, Kerou, M, Liu, H, Mcmahon, S, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-10 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4G
 
 | | Crystal structure of PA4631, a nucleoside-diphosphate-sugar epimerase from Pseudomonas aeruginosa | | Descriptor: | NUCLEOSIDE-DIPHOSPHATE-SUGAR EPIMERASE | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-30 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4I
 
 | | ORF 114a from Sulfolobus islandicus rudivirus 1 | | Descriptor: | UNCHARACTERIZED PROTEIN 114 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Naismith, J.H, White, M.F. | | Deposit date: | 2010-01-31 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X5P
 
 | | Crystal structure of the Streptococcus pyogenes fibronectin binding protein Fbab-B | | Descriptor: | FIBRONECTIN BINDING PROTEIN | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-10 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X3G
 
 | | Crystal Structure of the hypothetical protein ORF119 from Sulfolobus islandicus rod-shaped virus 1 | | Descriptor: | SIRV1 HYPOTHETICAL PROTEIN ORF119 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3M
 
 | | Crystal Structure of Hypothetical Protein ORF239 from Pyrobaculum Spherical Virus | | Descriptor: | HYPOTHETICAL PROTEIN ORF239 | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X4L
 
 | | Crystal structure of DesE, a ferric-siderophore receptor protein from Streptomyces coelicolor | | Descriptor: | FERRIC-SIDEROPHORE RECEPTOR PROTEIN | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X7I
 
 | | Crystal structure of mevalonate kinase from methicillin-resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, CITRIC ACID, MEVALONATE KINASE | | Authors: | Oke, M, Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-27 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
8PCW
 
 | | Structure of Csm6' from Streptococcus thermophilus | | Descriptor: | CRISPR system endoribonuclease Csm6' | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-11 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
8PE3
 
 | | Structure of Csm6' from Streptococcus thermophilus in complex with cyclic hexa-adenylate (cA6) | | Descriptor: | CRISPR system endoribonuclease Csm6', Cyclic hexaadenosine monophosphate (cA6), RNA | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-13 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
3FFE
 
 | | Structure of Achromobactin Synthetase Protein D, (AcsD) | | Descriptor: | AcsD | | Authors: | McMahon, S.A, Liu, H, Carter, L, Oke, M, Johnson, K.A, Schmelz, S, Challis, G.L, White, M.F, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2008-12-03 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
7BDV
 
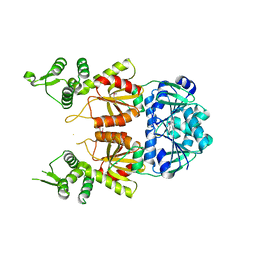 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
2MNA
 
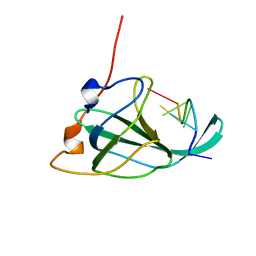 | | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus | | Descriptor: | Single-stranded DNA binding protein (SSB), ssDNA | | Authors: | Gamsjaeger, R, Kariawasam, R, Gimenez, A.X, Touma, C.F, McIlwain, E, Bernardo, R.E, Shepherd, N.E, Ataide, S.F, Dong, A.Q, Richard, D.J, White, M.F, Cubeddu, L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus
Biochem.J., 465, 2015
|
|
6SCE
 
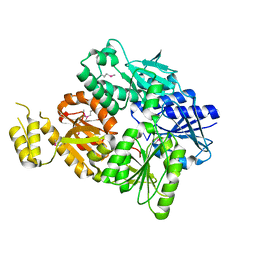 | | Structure of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Zhu, W, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate.
Nat Commun, 11, 2020
|
|
