1I5Z
 
 | | STRUCTURE OF CRP-CAMP AT 1.9 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | White, M.A, Lee, J.C, Fox, R.O. | | Deposit date: | 2001-03-01 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The effect of the D53H point mutation on the macroscopic
motions of E. coli Cyclic AMP Receptor Protein
To be Published
|
|
1I6X
 
 | | STRUCTURE OF A STAR MUTANT CRP-CAMP AT 2.2 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | White, M.A, Lee, J.C, Fox, R.O. | | Deposit date: | 2001-03-06 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The effect of the D53H point mutation on the macroscopic
motions of E. coli Cyclic AMP Receptor Protein
To be Published
|
|
7JQQ
 
 | | The bacteriophage Phi-29 viral genome packaging motor assembly | | Descriptor: | DNA (60-MER), DNA packaging protein, MAGNESIUM ION, ... | | Authors: | White, M.A, Woodson, M, Morais, M.C. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA.
Sci Adv, 7, 2021
|
|
3ZC4
 
 | | The structure of Csa5 from Sulfolobus solfataricus. | | Descriptor: | DI(HYDROXYETHYL)ETHER, SSO1398 | | Authors: | Reeks, J, Anderson, L, White, M.F, Naismith, J.H. | | Deposit date: | 2012-11-15 | | Release date: | 2013-02-20 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of the Archaeal Cascade Subunit Csa5: Relating the Small Subunits of Crispr Effector Complexes.
RNA Biol., 10, 2013
|
|
2W04
 
 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with citrate in ATP binding site from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, CITRATE ANION | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
3FFE
 
 | | Structure of Achromobactin Synthetase Protein D, (AcsD) | | Descriptor: | AcsD | | Authors: | McMahon, S.A, Liu, H, Carter, L, Oke, M, Johnson, K.A, Schmelz, S, Challis, G.L, White, M.F, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2008-12-03 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
1JTS
 
 | |
1JRE
 
 | | DNA PROTECTION AND BINDING BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, DNA PROTECTION DURING STARVATION PROTEIN | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2001-08-13 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA Protection and Binding by E. Coli Dps Protein
To be Published
|
|
4WNI
 
 | |
5NHQ
 
 | | Nuclear Magnetic Resonance Structure of the Human Polyoma JC Virus Agnoprotein | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, A.S, Abou-Gharbia, M, Childers, W, Condra, J, White, M.K, Safak, M, Bouaziz, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of the Human Polyoma JC Virus Agnoprotein.
J. Cell. Biochem., 118, 2017
|
|
2MJ2
 
 | | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, S.A, Abou-Gharbia, M, Childers, W, White, M, Bouaziz, S, Safak, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix
J.Virol., 2014
|
|
8PCW
 
 | | Structure of Csm6' from Streptococcus thermophilus | | Descriptor: | CRISPR system endoribonuclease Csm6' | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-11 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
8PE3
 
 | | Structure of Csm6' from Streptococcus thermophilus in complex with cyclic hexa-adenylate (cA6) | | Descriptor: | CRISPR system endoribonuclease Csm6', Cyclic hexaadenosine monophosphate (cA6), RNA | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-13 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
8BMW
 
 | | SsoCsm | | Descriptor: | CRISPR-associated Cas7 paralog (Type III-D), CRISPR-associated protein Cas10 (Type III-D), CRISPR-associated protein Cas5 (Type III-D), ... | | Authors: | Spagnolo, L, White, M.F. | | Deposit date: | 2022-11-11 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Saccharolobus solfataricus type III-D CRISPR effector.
Curr Res Struct Biol, 5, 2023
|
|
1EQV
 
 | |
1F2M
 
 | |
1F2Y
 
 | |
1F3W
 
 | | RECOMBINANT RABBIT MUSCLE PYRUVATE KINASE | | Descriptor: | MANGANESE (II) ION, POTASSIUM ION, PYRUVATE KINASE, ... | | Authors: | Wooll, J.O, Friesen, R.H.E, White, M.A, Watowich, S.J, Fox, R.O, Lee, J.C, Czerwinski, E.W. | | Deposit date: | 2000-06-06 | | Release date: | 2001-10-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional linkages between subunit interfaces in mammalian pyruvate kinase.
J.Mol.Biol., 312, 2001
|
|
1F3X
 
 | | S402P MUTANT OF RABBIT MUSCLE PYRUVATE KINASE | | Descriptor: | MANGANESE (II) ION, POTASSIUM ION, PYRUVATE KINASE, ... | | Authors: | Wooll, J.O, Friesen, R.H.E, White, M.A, Watowich, S.J, Fox, R.O, Lee, J.C, Czerwinski, E.W. | | Deposit date: | 2000-06-06 | | Release date: | 2001-10-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional linkages between subunit interfaces in mammalian pyruvate kinase.
J.Mol.Biol., 312, 2001
|
|
6YUD
 
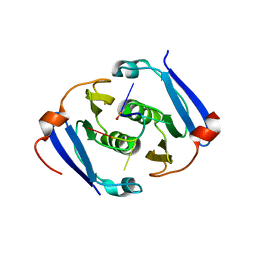 | | Structure of Csx3/Crn3 from Archaeoglobus fulgidus in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), Uncharacterized protein AF_1864 | | Authors: | McQuarrie, S, Gloster, T.M, White, M.F, Graham, S, Athukoralage, J.S, Gruschow, S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.
Elife, 9, 2020
|
|
7BDV
 
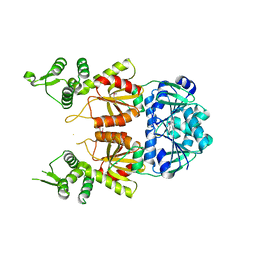 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
1F33
 
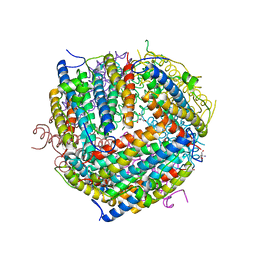 | |
1F30
 
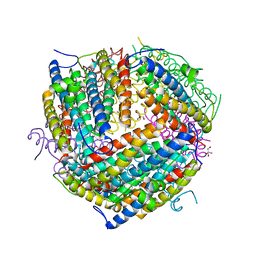 | | THE STRUCTURAL BASIS FOR DNA PROTECTION BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, ZINC ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2000-05-31 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structural Basis for DNA Protection by E. coli Dps Protein
To be Published
|
|
8B2X
 
 | |
2IVY
 
 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
