1AUL
 
 | | SOLUTION STRUCTURE OF A HIGHLY STABLE DNA DUPLEX CONJUGATED TO A MINOR GROOVE BINDER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*AP*AP*TP*CP*A)-3'), DNA (5'-D(P*THXP*GP*AP*TP*TP*AP*TP*CP*TP*G)-3') | | Authors: | Kumar, S, Reed, M.W, Gamper Junior, H.B, Gorn, V.V, Lukhtanov, E.A, Foti, M, West, J, Meyer Junior, R.B, Schweitzer, B.I. | | Deposit date: | 1997-08-29 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a highly stable DNA duplex conjugated to a minor groove binder.
Nucleic Acids Res., 26, 1998
|
|
1JBL
 
 | | Solution structure of SFTI-1, A cyclic trypsin inhibitor from sunflower seeds | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Korsinczky, M.L.J, Schirra, H.J, Rosengren, K.J, West, J, Condie, B.A, Otvos, L, Anderson, M.A, Craik, D.J. | | Deposit date: | 2001-06-05 | | Release date: | 2001-08-22 | | Last modified: | 2015-04-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant.
J.Mol.Biol., 311, 2001
|
|
1JBN
 
 | | Solution structure of an acyclic permutant of SFTI-1, A trypsin inhibitor from sunflower seeds | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Korsinczky, M.L.J, Schirra, H.J, Rosengren, K.J, West, J, Condie, B.A, Otvos, L, Anderson, M.A, Craik, D.J. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-22 | | Last modified: | 2016-12-28 | | Method: | SOLUTION NMR | | Cite: | Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant.
J.Mol.Biol., 311, 2001
|
|
1MI2
 
 | | SOLUTION STRUCTURE OF MURINE MACROPHAGE INFLAMMATORY PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN-2 | | Authors: | Shao, W, Jerva, L.F, West, J, Lolis, E, Schweitzer, B.I. | | Deposit date: | 1997-10-24 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of murine macrophage inflammatory protein-2.
Biochemistry, 37, 1998
|
|
6T38
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6TQG
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-12-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6T37
 
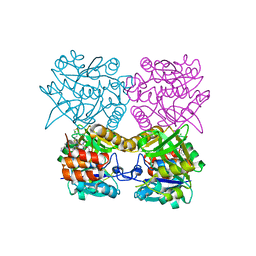 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
3HZX
 
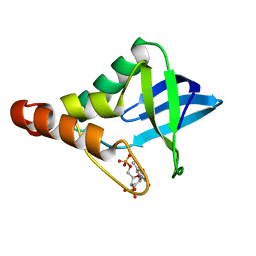 | |
2X4Z
 
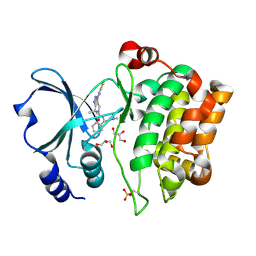 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with PF-03758309 | | Descriptor: | GLYCEROL, PF-3758309, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.R, Deng, Y, Murray, B, Guo, C, Piraino, J, Westwick, J, Zhang, C, Lamerdin, J, Dagostino, E, Loi, C.-M, Zager, M, Kraynov, E, Christensen, J, Martinez, R, Kephart, S, Marakovits, J, Karlicek, S, Bergqvist, S, Smeal, T. | | Deposit date: | 2010-02-03 | | Release date: | 2010-05-19 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-Molecule P21-Activated Kinase Inhibitor Pf- 3758309 is a Potent Inhibitor of Oncogenic Signaling and Tumor Growth.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1KCM
 
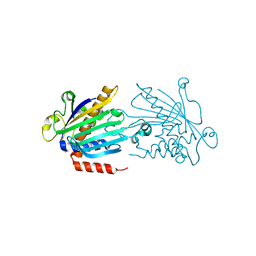 | | Crystal Structure of Mouse PITP Alpha Void of Bound Phospholipid at 2.0 Angstroms Resolution | | Descriptor: | Phosphatidylinositol Transfer Protein alpha | | Authors: | Schouten, A, Agianian, B, Westerman, J, Kroon, J, Wirtz, K.W.A, Gros, P. | | Deposit date: | 2001-11-09 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of apo-phosphatidylinositol transfer protein alpha provides insight into membrane association.
EMBO J., 21, 2002
|
|
3S2U
 
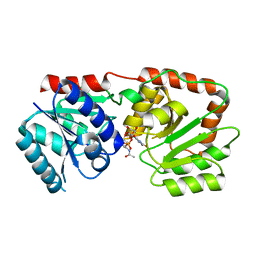 | | Crystal structure of the Pseudomonas aeruginosa MurG:UDP-GlcNAc substrate complex | | Descriptor: | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Brown, K, Vial, S.C.M, Dedi, N, Westcott, J, Scally, S, Bugg, T.D.H, Charlton, P.A, Cheetham, G.M.T. | | Deposit date: | 2011-05-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa MurG: UDP-GlcNAc Substrate Complex.
Protein Pept.Lett., 20, 2013
|
|
2AWN
 
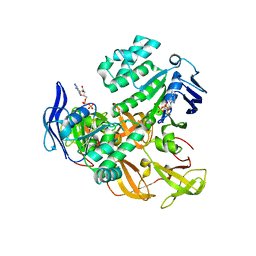 | | Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ATP-Mg) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Lu, G, Westbrooks, J.M, Davidson, A.L, Chen, J. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AWO
 
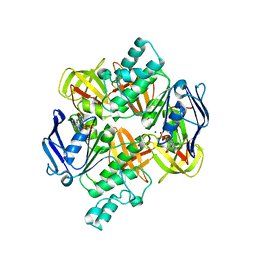 | | Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ADP-Mg) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Lu, G, Westbrooks, J.M, Davidson, A.L, Chen, J. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1QTS
 
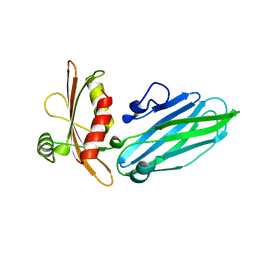 | |
1QTP
 
 | |
