1QGR
 
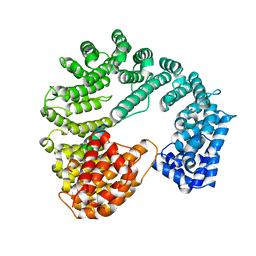 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA (II CRYSTAL FORM, GROWN AT LOW PH) | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-05-04 | | Release date: | 1999-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
8R6K
 
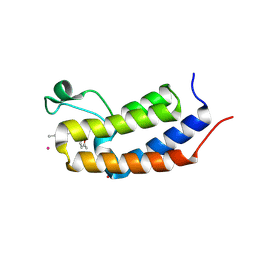 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a phenyltriazine ligand | | Descriptor: | 6-methyl-~{N}-[(5-methylfuran-2-yl)methyl]-3-(4-methylphenyl)-1,2,4-triazin-5-amine, Candida glabrata strain CBS138 chromosome C complete sequence, PHOSPHATE ION, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors With Antifungal Potential.
Adv Sci, 12, 2025
|
|
8R6L
 
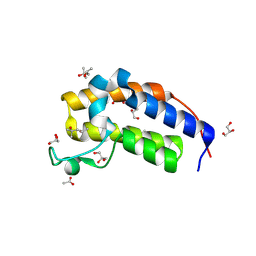 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 in the unbound state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Candida glabrata strain CBS138 chromosome C complete sequence, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors With Antifungal Potential.
Adv Sci, 12, 2025
|
|
8R6M
 
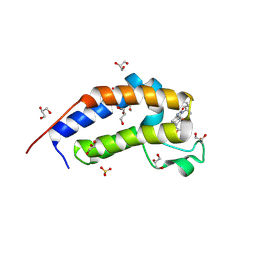 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 bound to I-BET151 | | Descriptor: | 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, Candida glabrata strain CBS138 chromosome C complete sequence, GLYCEROL, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors With Antifungal Potential.
Adv Sci, 12, 2025
|
|
8R6I
 
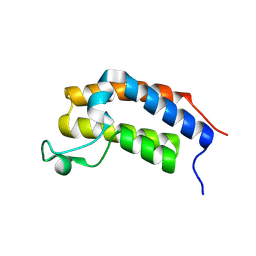 | |
8R6J
 
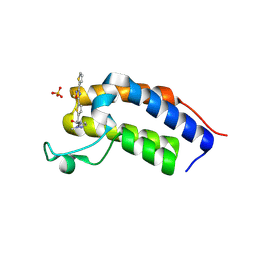 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a pyrazole ligand | | Descriptor: | 2-methyl-~{N}-[[5-(3-thiophen-2-yl-1,2,4-oxadiazol-5-yl)thiophen-2-yl]methyl]pyrazole-3-carboxamide, Candida glabrata strain CBS138 chromosome C complete sequence, SULFATE ION | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors With Antifungal Potential.
Adv Sci, 12, 2025
|
|
8R6N
 
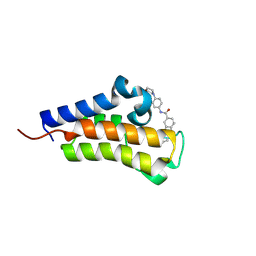 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 bound to a pyridoindole ligand | | Descriptor: | 2-ethanoyl-~{N}-(4-morpholin-4-ylphenyl)-1,3,4,5-tetrahydropyrido[4,3-b]indole-8-carboxamide, Candida glabrata strain CBS138 chromosome C complete sequence | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors With Antifungal Potential.
Adv Sci, 12, 2025
|
|
4DCQ
 
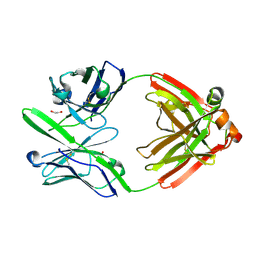 | | Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, 3B5H10 FAB Heavy Chain, 3B5H10 FAB Light Chain | | Authors: | Peters-Libeu, C.A, Tran, T, Finkbeiner, S, Weisgraber, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
6B85
 
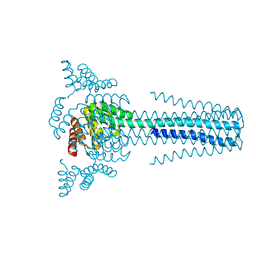 | | Crystal structure of transmembrane protein TMHC4_R | | Descriptor: | TMHC4_R | | Authors: | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.889 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6B87
 
 | | Crystal structure of transmembrane protein TMHC2_E | | Descriptor: | TMHC2_E | | Authors: | Lu, P, DiMaio, F, Min, D, Wei, K.Y, Bowie, J, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6BIT
 
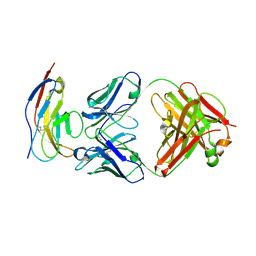 | | SIRPalpha antibody complex | | Descriptor: | KWAR23 Fab heavy chain, KWAR23 Fab light chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Ring, N.G, Herndler-Brandstetter, D, Weiskopf, K, Shan, L, Volkmer, J.P, George, B.M, Lietzenmayer, M, McKenna, K.M, Naik, T.J, McCarty, A, Zheng, Y, Ring, A.M, Flavell, R.A, Weissman, I.L. | | Deposit date: | 2017-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Anti-SIRP alpha antibody immunotherapy enhances neutrophil and macrophage antitumor activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6R9K
 
 | |
6R9L
 
 | |
8A7Z
 
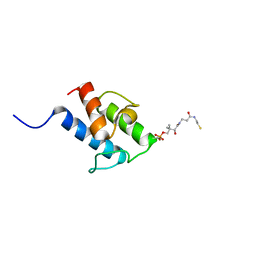 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AIG
 
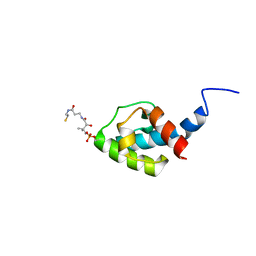 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8ALL
 
 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8RZX
 
 | |
3RYG
 
 | | 128 hours neutron structure of perdeuterated rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1KOA
 
 | | TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS | | Descriptor: | TWITCHIN | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
6ERL
 
 | |
8S81
 
 | | VirC mutant C114A-Q334A-R335A-R338A | | Descriptor: | 1,2-ETHANEDIOL, HMG-CoA synthase-like protein | | Authors: | Collin, S, Gruez, A, Weissman, K.J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Plasticity within 3-Hydroxy-3-Methylglutaryl Synthases Catalyzing the First Step of beta-Branching in Polyketide Biosynthesis Underpins a Dynamic Mechanism of Substrate Accommodation.
Jacs Au, 4, 2024
|
|
8S6L
 
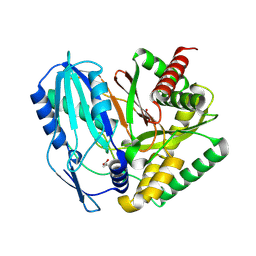 | | AzeD dehydratase of Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Phthiocerol/phthiodiolone dimycocerosyl transferase | | Authors: | Calderari, A, Gruez, A, Najah, S, Li, Y, Weissman, K.J. | | Deposit date: | 2024-02-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High resolution structure of azetidomonamide dehydratase
To be published
|
|
6V8Y
 
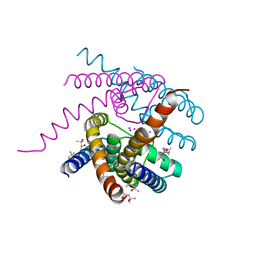 | | Structure of a Sodium Potassium ion Channel | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Roy, R, Weiss, K.L, Coates, L. | | Deposit date: | 2019-12-12 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural plasticity of the selectivity filter in a nonselective ion channel.
Iucrj, 8, 2021
|
|
8QKX
 
 | |
7PNE
 
 | |
