4BD1
 
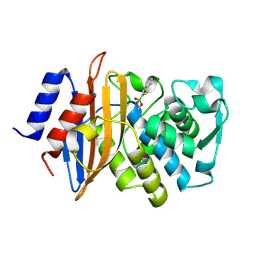 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-16 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.002 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
4BD0
 
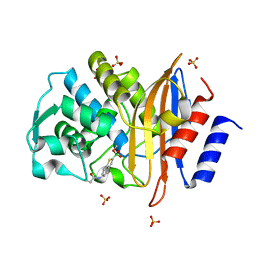 | | X-ray structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1, SULFATE ION | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
4GD6
 
 | | SHV-1 beta-lactamase in complex with penam sulfone SA1-204 | | Descriptor: | (3R)-N-(2-formylindolizin-3-yl)-4-[(phenylacetyl)oxy]-3-sulfino-D-valine, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | van den Akker, F, Wei, K. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structures of SHV-1 beta-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation
Plos One, 7, 2012
|
|
2JUG
 
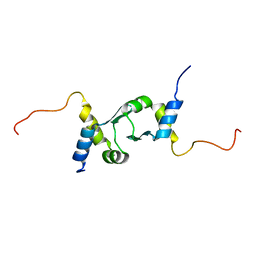 | |
2MF4
 
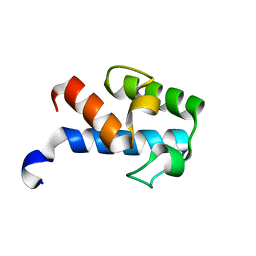 | | 1H, 13C, 15N chemical shift assignments of Streptomyces virginiae VirA acp5a | | Descriptor: | Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Davison, J, Dorival, J, Rabeharindranto, M.H, Mazon, H, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR assignements of ACP5a
To be Published
|
|
3RZ6
 
 | | Neutron structure of perdeuterated rubredoxin using 40 hours 1st pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RZT
 
 | | Neutron structure of perdeuterated rubredoxin using rapid (14 hours) data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.7504 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RYG
 
 | | 128 hours neutron structure of perdeuterated rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2KY7
 
 | | NMR Structural Studies on the Covalent DNA Binding of a Pyrrolobenzodiazepine-Naphthalimide Conjugate | | Descriptor: | 2-{2-[4-(3-{[(11aS)-7-methoxy-5-oxo-2,3,5,10,11,11a-hexahydro-1H-pyrrolo[2,1-c][1,4]benzodiazepin-8-yl]oxy}propyl)piperazin-1-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Langel, W, Kamal, A, Weisz, K. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on the covalent DNA binding of a pyrrolobenzodiazepine-naphthalimide conjugate
Org.Biomol.Chem., 8, 2010
|
|
3SS2
 
 | | Neutron structure of perdeuterated rubredoxin using 48 hours 3rd pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Blakeley, M.P, Weiss, K.L, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1YA9
 
 | | Crystal Structure of the 22kDa N-Terminal Fragment of Mouse Apolipoprotein E | | Descriptor: | Apolipoprotein E | | Authors: | Peters-Libeu, C.A, Rutenber, E, Newhouse, Y, Hatters, D.M, Weisgraber, K.H. | | Deposit date: | 2004-12-17 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Engineering conformational destabilization into mouse apolipoprotein E. A model for a unique property of human apolipoprotein E4
J.Biol.Chem., 280, 2005
|
|
2N5D
 
 | | NMR structure of PKS domains | | Descriptor: | fusion protein of two PKS domains | | Authors: | Dorival, J, Annaval, T, Risser, F, Collin, S, Roblin, P, Jacob, C, Gruez, A, Chagot, B, Weissman, K.J. | | Deposit date: | 2015-07-14 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Intersubunit Communication in the Virginiamycin trans-Acyl Transferase Polyketide Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
1IOJ
 
 | | HUMAN APOLIPOPROTEIN C-I, NMR, 18 STRUCTURES | | Descriptor: | APOC-I | | Authors: | Rozek, A, Sparrow, J.T, Weisgraber, K.H, Cushley, R.J. | | Deposit date: | 1998-05-12 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformation of human apolipoprotein C-I in a lipid-mimetic environment determined by CD and NMR spectroscopy.
Biochemistry, 38, 1999
|
|
1KOA
 
 | | TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS | | Descriptor: | TWITCHIN | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1KOB
 
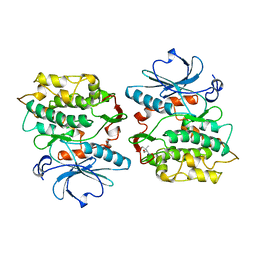 | | TWITCHIN KINASE FRAGMENT (APLYSIA), AUTOREGULATED PROTEIN KINASE DOMAIN | | Descriptor: | TWITCHIN, VALINE | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
5OV2
 
 | |
