4YZG
 
 | | Structure of the Arabidopsis TAP38/PPH1, a state-transition phosphatase responsible for dephosphorylation of LHCII | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2C 57, SULFATE ION | | Authors: | Wei, X.P, Guo, J.T, Li, M, Liu, Z.F. | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Mechanism Underlying the Specific Recognition between the Arabidopsis State-Transition Phosphatase TAP38/PPH1 and Phosphorylated Light-Harvesting Complex Protein Lhcb1
Plant Cell, 27, 2015
|
|
7RIG
 
 | | Structure of ACLY-D1026A-substrates | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-19 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7RKZ
 
 | | Structure of ACLY D1026A-substrates-asym-int | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-22 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7RMP
 
 | | Structure of ACLY D1026A - substrates-asym | | Descriptor: | (3S)-citryl-Coenzyme A, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-07-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7CTV
 
 | |
6KWN
 
 | | Crystal structure of pSLA-1*1301(F99Y) complex with S-OIV-derived epitope NSDTVGWSW | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
6KWO
 
 | | Crystal structure of pSLA-1*1301 complex with mutant epitope ESDTVGWSW | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
7CTX
 
 | |
6KWL
 
 | | Crystal structure of pSLA-1*0401(R156A) complex with FMDV-derived epitope MTAHITVPY | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
5LY0
 
 | | Crystal structure of LOB domain of Ramosa2 from Wheat | | Descriptor: | LOB family transfactor Ramosa2.1, ZINC ION | | Authors: | Wei, X.-B, Zhang, B, Chen, W.-F, Fan, S.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-23 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural analysis reveals a "molecular calipers" mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat.
J. Biol. Chem., 294, 2019
|
|
6UIA
 
 | |
6POE
 
 | | Structure of ACLY in complex with CoA | | Descriptor: | ATP-citrate synthase, COENZYME A | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8G1F
 
 | | Structure of ACLY-D1026A-products | | Descriptor: | (3S)-citryl-Coenzyme A, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
8G1E
 
 | | Structure of ACLY-D1026A-products-asym | | Descriptor: | (3S)-citryl-Coenzyme A, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
7SNI
 
 | | Structure of G6PD-D200N tetramer bound to NADP+ and G6P | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SNG
 
 | | structure of G6PD-WT tetramer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TOE
 
 | |
7TOF
 
 | | Structure of G6PD-WT dimer with no symmetry applied | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2022-01-24 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SNH
 
 | | Structure of G6PD-D200N tetramer bound to NADP+ | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SNF
 
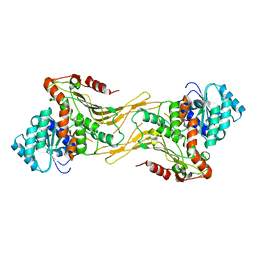 | | Structure of G6PD-WT dimer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UAL
 
 | |
7UC2
 
 | |
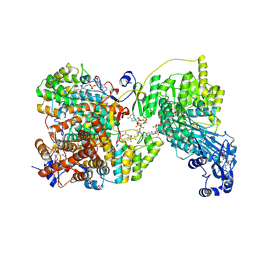
8G5D
 
 | |
4YZH
 
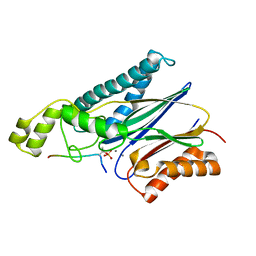 | | Structure of the Arabidopsis TAP38/PPH1 in complex with pLhcb1 phosphopeptide substrate | | Descriptor: | Chlorophyll a-b binding protein 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Wei, X.P, Guo, J.T, Li, M, Liu, Z.F. | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Mechanism Underlying the Specific Recognition between the Arabidopsis State-Transition Phosphatase TAP38/PPH1 and Phosphorylated Light-Harvesting Complex Protein Lhcb1
Plant Cell, 27, 2015
|
|
6UV5
 
 | |
