6Y96
 
 | | solution structure of cold-shock domain 9 of drosophila Upstream of N-Ras (Unr) | | Descriptor: | Upstream of N-ras, isoform A | | Authors: | Sweetapple, L.J, Hollmann, N.M, Simon, B, Hennig, J. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep, 32, 2020
|
|
8FIZ
 
 | | Cryo-EM structure of E. coli 70S Ribosome containing mRNA and tRNA (in the transcription-translation complex) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8FIY
 
 | | Cryo-EM structure of E. coli RNA polymerase Elongation complex in the Transcription-Translation Complex (RNAP in an anti-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8FIX
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex harboring a terminal mismatch | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
3L9I
 
 | | Myosin VI nucleotide-free (mdinsert2) L310G mutant crystal structure | | Descriptor: | ACETATE ION, CALCIUM ION, Calmodulin, ... | | Authors: | Pylypenko, O, Song, L, Sweeney, L.H, Houdusse, A. | | Deposit date: | 2010-01-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of insert i of myosin VI in modulating nucleotide affinity
To be Published
|
|
4ZG4
 
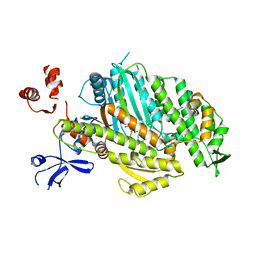 | | Myosin Vc Pre-powerstroke | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ropars, V, Pylypenko, O, Sweeney, L, Houdusse, A. | | Deposit date: | 2015-04-22 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Force-producing ADP state of myosin bound to actin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KG8
 
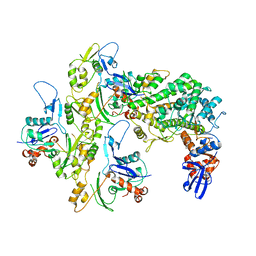 | |
2YE0
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
2YDZ
 
 | | X-ray structure of the cyan fluorescent protein SCFP3A (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
3ZTF
 
 | | X-ray Structure of the Cyan Fluorescent Protein mTurquoise2 (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
