7RFQ
 
 | |
6B7L
 
 | | Aeromonas veronii immune modulator A | | Descriptor: | CHLORIDE ION, FORMIC ACID, immune modulator A | | Authors: | Sweeney, E.S, Remington, S.J, Perkins, A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-03 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A bacterial immunomodulatory protein with lipocalin-like domains facilitates host-bacteria mutualism in larval zebrafish.
Elife, 7, 2018
|
|
7K5N
 
 | |
4EXO
 
 | | Revised, rerefined crystal structure of PDB entry 2QHK, methyl accepting chemotaxis protein | | Descriptor: | Methyl-accepting chemotaxis protein, PYRUVIC ACID | | Authors: | Sweeney, E.G, Henderson, J.N, Goers, J, Wreden, C, Hicks, K.G, Foster, J.K, Parthasarathy, R, Remington, S.J, Guillemin, K. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Proposed Mechanism for the pH-Sensing Helicobacter pylori Chemoreceptor TlpB.
Structure, 20, 2012
|
|
6VQ2
 
 | | HLA-B*27:05 presenting an HIV-1 14mer peptide | | Descriptor: | 14-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQE
 
 | | HLA-B*27:05 presenting an HIV-1 13mer peptide | | Descriptor: | 13-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQD
 
 | | HLA-B*27:05 presenting an HIV-1 8mer peptide | | Descriptor: | 8-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQY
 
 | | HLA-B*27:05 presenting an HIV-1 7mer peptide | | Descriptor: | 7-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQZ
 
 | | HLA-B*27:05 presenting an HIV-1 6mer peptide | | Descriptor: | 6-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VPZ
 
 | | HLA-B*27:05 presenting an HIV-1 11mer peptide | | Descriptor: | 11-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
5VJH
 
 | | Closed State CryoEM Reconstruction of Hsp104:ATPyS and FITC casein | | Descriptor: | FITC casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
6DTM
 
 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein TlpA | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-06-17 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
5VYA
 
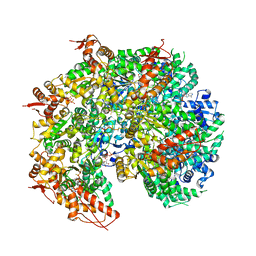 | | S. cerevisiae Hsp104:casein complex, Extended Conformation | | Descriptor: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VY8
 
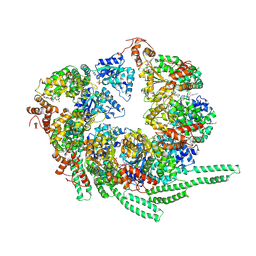 | | S. cerevisiae Hsp104-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 104 | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VY9
 
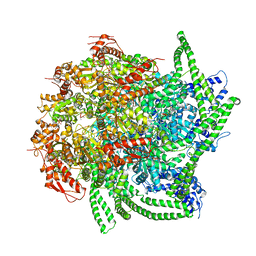 | | S. cerevisiae Hsp104:casein complex, Middle Domain Conformation | | Descriptor: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
6E0A
 
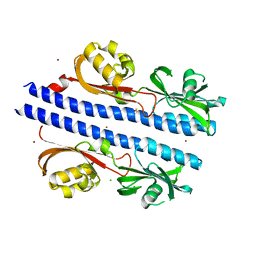 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-07-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
6E09
 
 | |
3UB7
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with acetamide bound | | Descriptor: | ACETAMIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3UB8
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with formamide bound | | Descriptor: | FORMAMIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3UB9
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with hydroxyurea bound | | Descriptor: | GLYCEROL, N-HYDROXYUREA, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3UB6
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with urea bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3QEB
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with DNA and Mn2+ (complex III) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QE9
 
 | | Crystal structure of human exonuclease 1 Exo1 (D173A) in complex with DNA (complex I) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QEA
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with DNA (complex II) | | Descriptor: | BARIUM ION, DNA (5'-D(P*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
