2A43
 
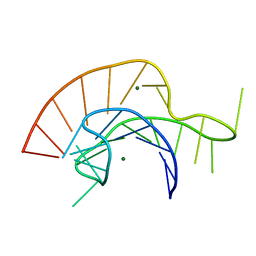 | | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif | | Descriptor: | MAGNESIUM ION, RNA Pseudoknot | | Authors: | Pallan, P.S, Marshall, W.S, Harp, J, Jewett III, F.C, Wawrzak, Z, Brown II, B.A, Rich, A, Egli, M. | | Deposit date: | 2005-06-27 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif
Biochemistry, 44, 2005
|
|
5IQJ
 
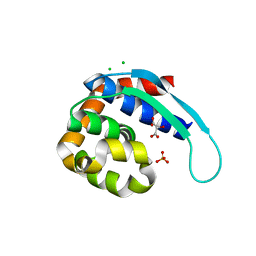 | | 1.9 Angstrom Crystal Structure of Protein with Unknown Function from Vibrio cholerae. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Protein with Unknown Function from Vibrio cholerae.
To Be Published
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
5CRF
 
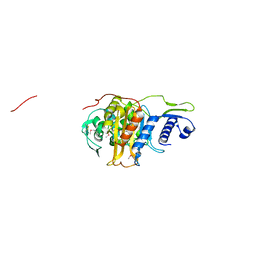 | | Structure of the penicillin-binding protein PonA1 from Mycobacterium Tuberculosis | | Descriptor: | PHOSPHATE ION, Penicillin-binding protein 1A | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-22 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5D6A
 
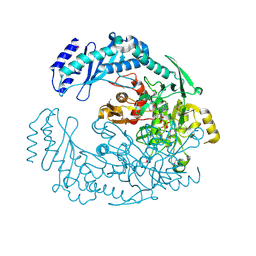 | | 2.7 Angstrom Crystal Structure of ABC transporter ATPase from Vibrio vulnificus in Complex with Adenylyl-imidodiphosphate (AMP-PNP) | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Predicted ATPase of the ABC class, SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom Crystal Structure of ABC transporter ATPase from Vibrio vulnificus in Complex with Adenylyl-imidodiphosphate (AMP-PNP)
To Be Published
|
|
1TP5
 
 | | Crystal structure of PDZ3 domain of PSD-95 protein complexed with a peptide ligand KKETWV | | Descriptor: | LYS-LYS-GLU-THR-TRP-VAL peptide ligand, Presynaptic density protein 95 | | Authors: | Saro, D, Wawrzak, Z, Martin, P, Vickrey, J, Paredes, A, Kovari, L, Spaller, M. | | Deposit date: | 2004-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of the third PDZ domain of PSD-95 protein complexed with KKETWV peptide ligand
To be Published
|
|
4Z8Z
 
 | | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Endres, M, Joachimiak, J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405
To Be Published
|
|
4ZV9
 
 | | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-18 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
1PNV
 
 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and Vancomycin | | Descriptor: | GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, VANCOMYCIN, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-13 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PWF
 
 | | One Sugar Pucker Fits All: Pairing Versatility Despite Conformational Uniformity in TNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2MU)P*(FA2)P*CP*GP*C)-3', SPERMINE | | Authors: | Pallan, P.S, Wilds, C.J, Wawrzak, Z, Krishnamurthy, R, Eschenmoser, A, Egli, M. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Why does TNA cross-pair more strongly with RNA than with DNA? an answer from X-ray analysis.
ANGEW.CHEM.INT.ED.ENGL., 42, 2003
|
|
1PN3
 
 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and the acceptor substrate DVV. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-12 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1RPI
 
 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | alpha-D-glucopyranose, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-03 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1RV7
 
 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-12 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
1RQ9
 
 | | Crystal structures of a Multidrug-Resistant HIV-1 Protease Reveal an Expanded Active Site Cavity | | Descriptor: | [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE, protease | | Authors: | Logsdon, B.C, Vickrey, J.F, Martin, P, Proteasa, G, Koepke, J.I, Terlecky, S.R, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | Deposit date: | 2003-12-04 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity.
J.Virol., 78, 2004
|
|
6N7M
 
 | | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630. | | Descriptor: | Hypothetical Protein CD630_05490 | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630.
To Be Published
|
|
6NFP
 
 | | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | 1,2-ETHANEDIOL, Arginase, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
6MCW
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, complex with the detergent Anapoe-X-114 | | Descriptor: | 23-[4-(2,4,4-trimethylpentan-2-yl)phenoxy]-3,6,9,12,15,18,21-heptaoxatricosan-1-ol, Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-02 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
6MI0
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, ligand-free state | | Descriptor: | Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-18 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
6N0I
 
 | | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongation factor G 2, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Elongation Factor G 2 from Pseudomonas putida.
To Be Published
|
|
1I0T
 
 | | 0.6 A STRUCTURE OF Z-DNA CGCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', SPERMINE | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.6 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
6NDI
 
 | | Crystal Structure of the Sugar Binding Domain of LacI Family Protein from Klebsiella pneumoniae | | Descriptor: | Transcriptional regulator | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-13 | | Release date: | 2018-12-26 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
3UUW
 
 | | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.
TO BE PUBLISHED
|
|
3T4X
 
 | | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. Ames Ancestor | | Descriptor: | Oxidoreductase, short chain dehydrogenase/reductase family | | Authors: | Filippova, E.V, Wawrzak, Z, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3SS6
 
 | | Crystal structure of the Bacillus anthracis acetyl-CoA acetyltransferase | | Descriptor: | Acetyl-CoA acetyltransferase, POTASSIUM ION, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Bacillus anthracis acetyl-CoA acetyltransferase
TO BE PUBLISHED
|
|
3TYS
 
 | | Crystal structure of transcriptional regulator VanUg, Form II | | Descriptor: | Predicted transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form II
TO BE PUBLISHED
|
|
