8GSR
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (apo-form) | | 分子名称: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION | | 著者 | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | 登録日 | 2022-09-07 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
5GN9
 
 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17b | | 分子名称: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | 著者 | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | 登録日 | 2016-07-19 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5GN5
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17 | | 分子名称: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | 著者 | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | 登録日 | 2016-07-19 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
7CNQ
 
 | | Crystal structure of Agrobacterium tumefaciens aconitase X (holo-form) | | 分子名称: | (2~{S},3~{R})-3-oxidanylpyrrolidine-2-carboxylic acid, FE2/S2 (INORGANIC) CLUSTER, cis-3-hydroxy-L-proline dehydratase | | 著者 | Murase, Y, Watanabe, Y, Watanabe, S. | | 登録日 | 2020-08-03 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7CNR
 
 | |
7CNP
 
 | |
7CNS
 
 | | Crystal structure of Thermococcus kodakaraensis aconitase X (holo-form) | | 分子名称: | (3R)-3-HYDROXY-3-METHYL-5-(PHOSPHONOOXY)PENTANOIC ACID, DUF521 domain-containing protein, FE3-S4 CLUSTER, ... | | 著者 | Murase, Y, Watanabe, Y, Watanabe, S. | | 登録日 | 2020-08-03 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7YCF
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis IN ACETONITRILE | | 分子名称: | 2-HYDROXY-2-METHYLPROPANENITRILE, CHLORIDE ION, Hydroxynitrile lyase, ... | | 著者 | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | 登録日 | 2022-07-01 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCB
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE | | 分子名称: | CHLORIDE ION, GLYCEROL, Hydroxynitrile lyase, ... | | 著者 | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | 登録日 | 2022-07-01 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCT
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis complexed with (R)-2-Chloromandelonitrile | | 分子名称: | (2~{R})-2-(2-chlorophenyl)-2-oxidanyl-ethanenitrile, GLYCEROL, Hydroxynitrile lyase, ... | | 著者 | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | 登録日 | 2022-07-01 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCD
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis bound with (R)-(+)-ALPHA-HYDROXYBENZENE-ACETONITRILE | | 分子名称: | (2R)-hydroxy(phenyl)ethanenitrile, Hydroxynitrile lyase, SULFATE ION | | 著者 | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | 登録日 | 2022-07-01 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YAX
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, | | 分子名称: | CHLORIDE ION, Hydroxynitrile lyase, SULFATE ION | | 著者 | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | 登録日 | 2022-06-28 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
2NLI
 
 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | 分子名称: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | 著者 | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | 登録日 | 2006-10-20 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
7CGQ
 
 | |
7DO6
 
 | |
7B81
 
 | |
7CGR
 
 | |
5GN6
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17b | | 分子名称: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | 著者 | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | 登録日 | 2016-07-19 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
1D8K
 
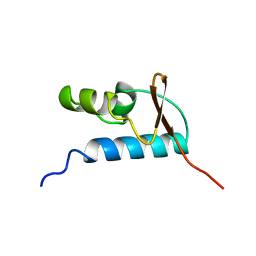 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | 分子名称: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | 著者 | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1999-10-25 | | 公開日 | 2000-04-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
5GN7
 
 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17 | | 分子名称: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | 著者 | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | 登録日 | 2016-07-19 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
7D2R
 
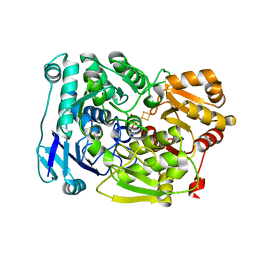 | | Crystal structure of Agrobacterium tumefaciens aconitase X mutant - S449C/C510V | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, SODIUM ION, ... | | 著者 | Murase, Y, Watanabe, Y, Watanabe, S. | | 登録日 | 2020-09-17 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7DO5
 
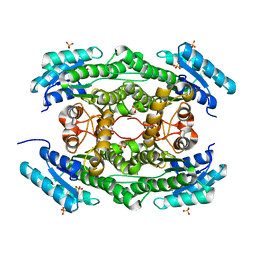 | |
7DO7
 
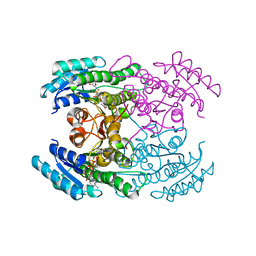 | |
7FGP
 
 | |
1D8J
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | 分子名称: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | 著者 | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1999-10-25 | | 公開日 | 2000-04-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
