4LLS
 
 | | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with IPP, GSPP, and calcium bound in active site | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with IPP, GSPP and calcium bound in active site
To be Published
|
|
4MK3
 
 | | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h) | | Descriptor: | Glutathione S-transferase, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, MAGNESIUM ION, ... | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h)
To be published
|
|
4RY8
 
 | | Crystal structure of 5-methylthioribose transporter solute binding protein TLET_1677 from Thermotoga lettingae TMO TARGET EFI-511109 in complex with 5-methylthioribose | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, Periplasmic binding protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of 5-methylthioribose binding protein TLET_1677 from Thermotoga lettingae TARGET EFI-511109
To be Published
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
4H83
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing enzyme (EFI target:502127) | | Descriptor: | BICARBONATE ION, GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing enzyme
To be Published
|
|
4HI7
 
 | | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione | | Descriptor: | GI20122, GLUTATHIONE | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-11 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione
To be Published
|
|
4HOJ
 
 | | Crystal structure of glutathione transferase homolog from Neisseria Gonorrhoeae, target EFI-501841, with bound glutathione | | Descriptor: | ACETATE ION, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of glutathione transferase homolog from Neisseria Gonorrhoeae, target EFI-501841, with bound glutathione
To be Published
|
|
4ID0
 
 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione sulfinic acid (gso2h) and acetate | | Descriptor: | ACETATE ION, GLYCEROL, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-11 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione sulfinic acid (gso2h) and acetate
To be Published
|
|
4IGJ
 
 | | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, target EFI-507175 | | Descriptor: | Maleylacetoacetate isomerase, SULFATE ION | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, target EFI-507175
To be Published
|
|
4IKH
 
 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-26 | | Release date: | 2013-01-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound
To be Published
|
|
4XFM
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, with bound D-threonate, domain swapped dimer | | Descriptor: | THREONATE ION, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-27 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ISD
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE homolog from BURKHOLDERIA GL BGR1, TARGET EFI-501803, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of GLUTATHIONE TRANSFERASE homolog from BURKHOLDERIA GL BGR1, TARGET EFI-501803, with bound glutathione
TO BE PUBLISHED
|
|
4J2F
 
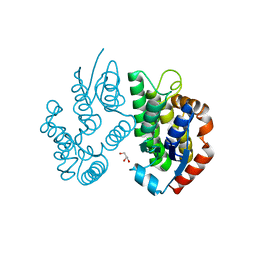 | | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866 | | Descriptor: | GLYCEROL, Glutathione s-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-02-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866
TO BE PUBLISHED
|
|
4IVF
 
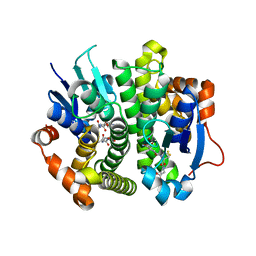 | | Crystal structure of glutathione transferase homolog from Lodderomyces elongisporus, target EFI-501753, with two GSH per subunit | | Descriptor: | CITRIC ACID, GLUTATHIONE, Putative uncharacterized protein | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-22 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione transferase homolog from Lodderomyces elongisporus, target EFI-501753, with two GSH per subunit
To be Published
|
|
4XG0
 
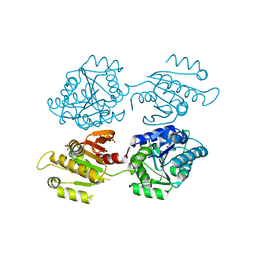 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group C2221 | | Descriptor: | CHLORIDE ION, CITRIC ACID, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4JCI
 
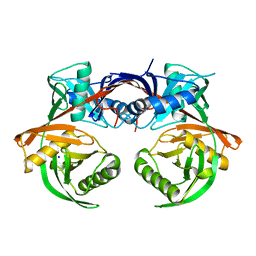 | | Crystal structure of csal_2705, a putative hydroxyproline epimerase from CHROMOHALOBACTER SALEXIGENS (TARGET EFI-506486), SPACE GROUP P212121, unliganded | | Descriptor: | Proline racemase, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of csal_2705, a putative hydroxyproline epimerase from CHROMOHALOBACTER SALEXIGENS (TARGET EFI-506486), space group P212121, unliganded
To be Published
|
|
4J9W
 
 | | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with the inhibitor pyrrole-2-carboxylate | | Descriptor: | GLYCEROL, PYRROLE-2-CARBOXYLATE, Proline racemase family protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with the inhibitor pyrrole-2-carboxylate
To be Published
|
|
4JBD
 
 | | Crystal structure of Pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group I2, bound citrate | | Descriptor: | CITRIC ACID, Proline racemase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from pseudomonas putida f1 (target efi-506500), open form, space group i2, bound citrate
To be Published
|
|
4JUU
 
 | | Crystal structure of a putative hydroxyproline epimerase from xanthomonas campestris (TARGET EFI-506516) with bound phosphate and unknown ligand | | Descriptor: | CHLORIDE ION, Epimerase, PHOSPHATE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative hydroxyproline epimerase from xanthomonas campestris (TARGET EFI-506516) with bound phosphate and unknown ligand
To be Published
|
|
4JD7
 
 | | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate | | Descriptor: | Proline racemase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate
To be Published
|
|
2QSR
 
 | | Crystal structure of C-terminal domain of transcription-repair coupling factor | | Descriptor: | Transcription-repair coupling factor | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Bain, K, Iizuka, M, Wasserman, S, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of C-terminal domain of transcription-repair coupling factor.
To be Published
|
|
4J9X
 
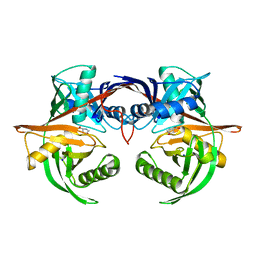 | | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline | | Descriptor: | 4-HYDROXYPROLINE, Proline racemase family protein, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline
To be Published
|
|
4JN8
 
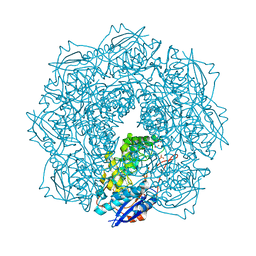 | | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ENOLASE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site
To be Published
|
|
4JN7
 
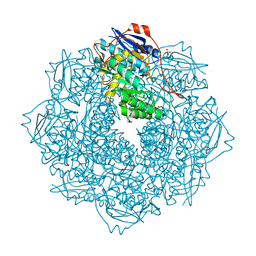 | | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE
To be Published
|
|
4PTS
 
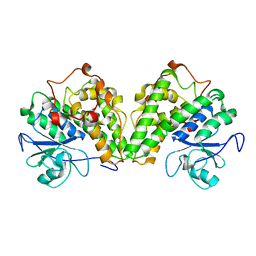 | | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405 | | Descriptor: | glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405
To be Published
|
|
