2J2S
 
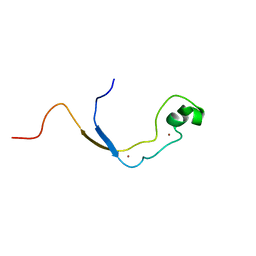 | | Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase | | Descriptor: | ZINC FINGER PROTEIN HRX, ZINC ION | | Authors: | Allen, M.D, Grummitt, C.G, Hilcenko, C, Young-Min, S, Tonkin, L.M, Johnson, C.M, Bycroft, M, Warren, A.J. | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Nonmethyl-Cpg-Binding Cxxc Domain of the Leukaemia-Associated Mll Histone Methyltransferase
Embo J., 25, 2006
|
|
1O8P
 
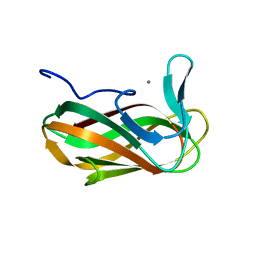 | | Unbound structure of CsCBM6-3 from Clostridium stercorarium | | Descriptor: | CALCIUM ION, PUTATUVE ENDO-XYLANASE | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains.
J. Mol. Biol., 327, 2003
|
|
2HIS
 
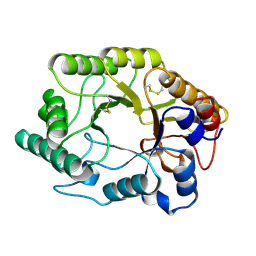 | | CELLULOMONAS FIMI XYLANASE/CELLULASE DOUBLE MUTANT E127A/H205N WITH COVALENT CELLOBIOSE | | Descriptor: | CELLULOMONAS FIMI FAMILY 10 BETA-1,4-GLYCANASE, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Notenboom, V, Birsan, C, Nitz, M, Rose, D.R, Warren, R.A.J, Wither, S.G. | | Deposit date: | 1998-02-23 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insights into transition state stabilization of the beta-1,4-glycosidase Cex by covalent intermediate accumulation in active site mutants.
Nat.Struct.Biol., 5, 1998
|
|
1NAE
 
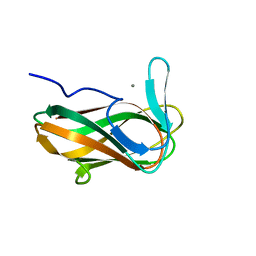 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with xylotriose | | Descriptor: | CALCIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, putative xylanase | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains
J.Mol.Biol., 327, 2003
|
|
2XYL
 
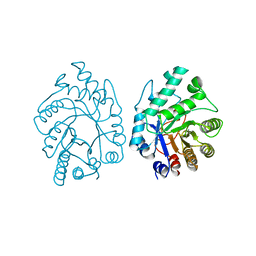 | | CELLULOMONAS FIMI XYLANASE/CELLULASE COMPLEXED WITH 2-DEOXY-2-FLUORO-XYLOBIOSE | | Descriptor: | BETA-1,4-GLYCANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Monem, V, Birsan, C, Warren, R.A.J, Withers, S.G, Rose, D.R. | | Deposit date: | 1997-11-20 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the cellulose/xylan specificity of the beta-1,4-glycanase cex from Cellulomonas fimi through crystallography and mutation.
Biochemistry, 37, 1998
|
|
1ODZ
 
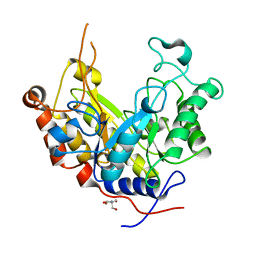 | | Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mannan endo-1,4-beta-mannosidase, SODIUM ION, ... | | Authors: | Jahn, M, Stoll, D, Warren, R.A.J, Szabo, L, Singh, P, Gilbert, H.J, Ducros, V.M.A, Davies, G.J, Withers, S.G. | | Deposit date: | 2003-03-17 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expansion of the Glycosynthase Repertoire to Produce Defined Manno-Oligosaccharides
Chem.Commun.(Camb.), 12, 2003
|
|
1O8S
 
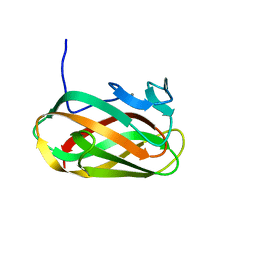 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with cellobiose | | Descriptor: | CALCIUM ION, PUTATIVE ENDO-XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
1OD3
 
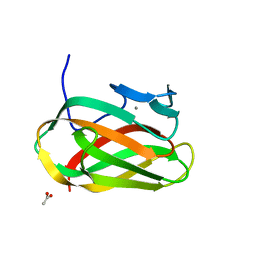 | | Structure of CSCBM6-3 From Clostridium stercorarium in complex with laminaribiose | | Descriptor: | ACETIC ACID, CALCIUM ION, PUTATIVE XYLANASE, ... | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
2JF4
 
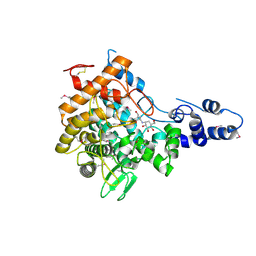 | | Family 37 trehalase from Escherichia coli in complex with validoxylamine | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, PERIPLASMIC TREHALASE | | Authors: | Gibson, R.P, Gloster, T.M, Roberts, S, Warren, R.A.J, Storch De Gracia, I, Garcia, A, Chiara, J.L, Davies, G.J. | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for Trehalase Inhibition Revealed by the Structure of Trehalase in Complex with Potent Inhibitors.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
1I82
 
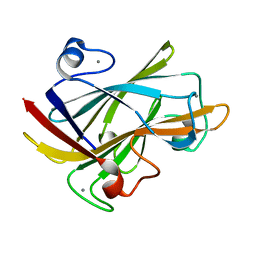 | | FAMILY 9 CARBOHYDRATE-BINDING MODULE FROM THERMOTOGA MARITIMA XYLANASE 10A WITH CELLOBIOSE | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Warren, R.A.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-03-12 | | Release date: | 2001-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms.
Biochemistry, 40, 2001
|
|
1I8U
 
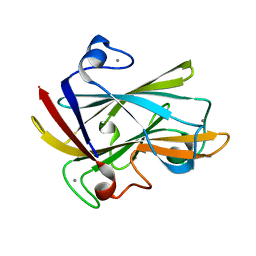 | | FAMILY 9 CARBOHYDRATE-BINDING MODULE FROM THERMOTOGA MARITIMA XYLANASE 10A | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A | | Authors: | Notenboom, V, Boraston, A.B, Warren, R.A.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-03-16 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms.
Biochemistry, 40, 2001
|
|
1I8A
 
 | | FAMILY 9 CARBOHYDRATE-BINDING MODULE FROM THERMOTOGA MARITIMA XYLANASE 10A WITH GLUCOSE | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Warren, R.A.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-03-12 | | Release date: | 2001-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms.
Biochemistry, 40, 2001
|
|
2JG0
 
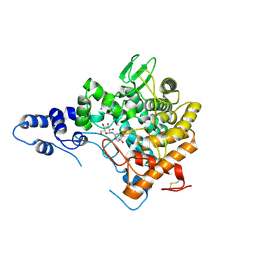 | | Family 37 trehalase from Escherichia coli in complex with 1- thiatrehazolin | | Descriptor: | N-[(3aS,4R,5S,6S,6aS)-4,5,6-trihydroxy-4-(hydroxymethyl)-4,5,6,6a-tetrahydro-3aH-cyclopenta[d][1,3]thiazol-2-yl]-alpha- D-glucopyranosylamine, PERIPLASMIC TREHALASE | | Authors: | Gibson, R.P, Gloster, T.M, Roberts, S, Warren, R.A.J, Storch De Gracia, I, Garcia, A, Chiara, J.L, Davies, G.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis for Trehalase Inhibition Revealed by the Structure of Trehalase in Complex with Potent Inhibitors.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
