7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPD
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with one JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPF
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with three JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WRV
 
 | | The interface of JMB2002 Fab binds to SARS-CoV-2 Omicron Variant S | | Descriptor: | JMB2002 Fab heavy chain, JMB2002 Fab light chain, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPE
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with two JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
7V9L
 
 | | Cryo-EM structure of the SV1-Gs complex. | | Descriptor: | GHRH receptor splice variant 1,GHRH receptor splice variant 1,GHRH receptor splice variant 1,SV1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Cheng, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8HKD
 
 | |
7DUQ
 
 | | Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7DUR
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7E5S
 
 | | SARS-CoV-2 S trimer with four-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
7EVM
 
 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
6DII
 
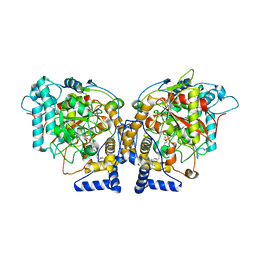 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase in Complex with methyl linolenyl fluorophosphonate | | Descriptor: | Fatty acid amide hydrolase, methyl-9Z,12Z,15Z-octadecatrienylphosphonofluoridate | | Authors: | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | Deposit date: | 2018-05-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
7F3Q
 
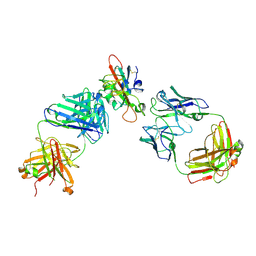 | | SARS-CoV-2 RBD in complex with A5-10 Fab and A34-2 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A34-2 Fab, ... | | Authors: | Dou, Y, Wang, X, Liu, P, Lu, B, Wang, K. | | Deposit date: | 2021-06-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Development of neutralizing antibodies against SARS-CoV-2, using a high-throughput single-B-cell cloning method.
Antib Ther, 6, 2023
|
|
6DHV
 
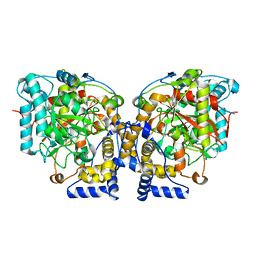 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
2KCW
 
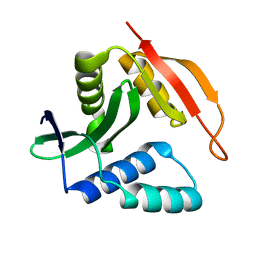 | |
2NC5
 
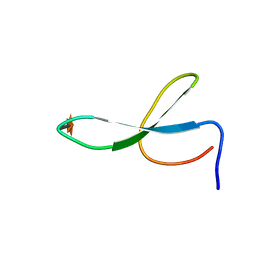 | | Solution Structure of N-Xylosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-xylopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
7E5R
 
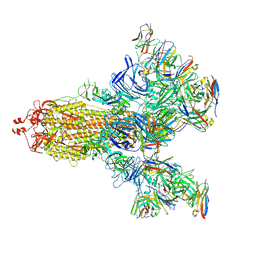 | | SARS-CoV-2 S trimer with three-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
5ZMY
 
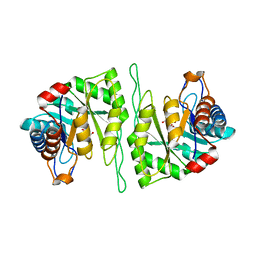 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, D(-)-TARTARIC ACID, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
5ZMU
 
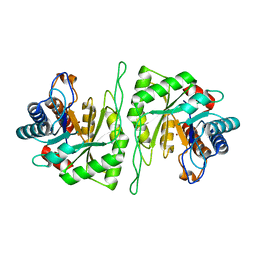 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
6IUV
 
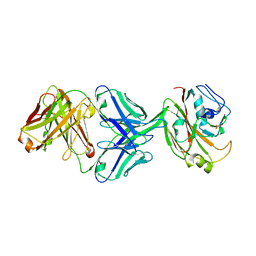 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 3C11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3C11 Heavy Chain, 3C11 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Structural and functional definition of a vulnerable site on the hemagglutinin of highly pathogenic avian influenza A virus H5N1.
J. Biol. Chem., 294, 2019
|
|
7D7N
 
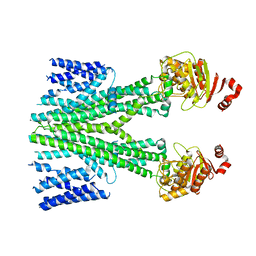 | | Cryo-EM structure of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
7D7R
 
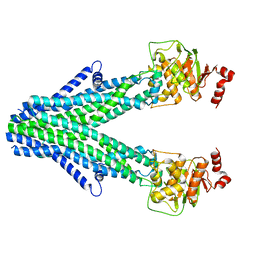 | | Cryo-EM structure of the core domain of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
