7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7EZP
 
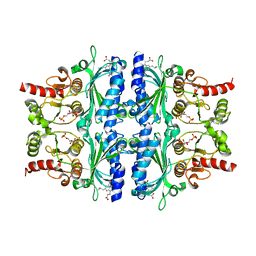 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
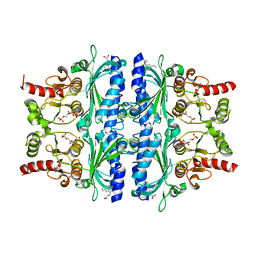 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
5ZT3
 
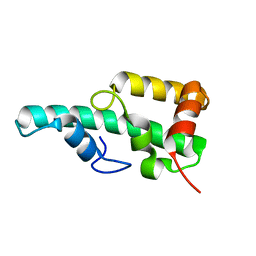 | | Crystal structure of WA352 from Oryza sativa | | Descriptor: | WA352 | | Authors: | Wang, X, Guan, Z, Yin, P. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Crystal structure of WA352 provides insight into cytoplasmic male sterility in rice
Biochem. Biophys. Res. Commun., 501, 2018
|
|
6AJ6
 
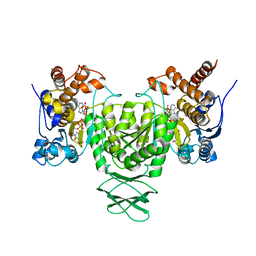 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADP+ | | Descriptor: | Isocitrate dehydrogenase [NADP], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
2GYZ
 
 | | Crystal structure of human artemin | | Descriptor: | neurotrophic factor artemin isoform 3 | | Authors: | Wang, X.Q, Garcia, K.C. | | Deposit date: | 2006-05-10 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
2GH0
 
 | | Growth factor/receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDNF family receptor alpha-3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
2GYR
 
 | | Crystal structure of human artemin | | Descriptor: | Neurotrophic factor artemin, isoform 3 | | Authors: | Wang, X.Q. | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
6L77
 
 | | Crystal structure of MS5 from Brassica napus | | Descriptor: | MS5a | | Authors: | Wang, X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the meiosis-related protein MS5 reveals non-canonical papain enhancement by cystatin-like folds.
Febs Lett., 594, 2020
|
|
7EGR
 
 | | Co-crystal structure of Ac-AChBPP in complex with RgIA | | Descriptor: | MAGNESIUM ION, RgIA, Soluble acetylcholine receptor | | Authors: | Wang, X.Q, Pan, S, Fan, Y.X, Xue, Y, Zhu, X.P, Luo, S.L. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Co-crystal structure of Ac-AChBPP in complex with RgIA
To Be Published
|
|
7FC6
 
 | | Crystal structure of SARS-CoV RBD and horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, X.Q, Lan, J, Ge, J.W. | | Deposit date: | 2021-07-13 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
6IZF
 
 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | (2R)-1-(dodecanoyloxy)-3-hydroxypropan-2-yl (5E,8E,11E)-tetradeca-5,8,11-trienoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, X.H, Zeng, Y, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7BIK
 
 | | Crystal structure of YTHDF2 in complex with m6Am | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(hydroxymethyl)-4-methoxy-5-[6-(methylamino)purin-9-yl]oxolan-3-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Wang, X, Caflisch, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YTHDF2 in complex with m6Am
To Be Published
|
|
6IYZ
 
 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Qin, L, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IYU
 
 | | Crystal Structure Analysis of an Eukaryotic Membrane Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Su, M, Gao, F, Chen, Y.H. | | Deposit date: | 2018-12-17 | | Release date: | 2019-05-01 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ1
 
 | | Crystal Structure Analysis of a Eukaryotic Membrane Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ0
 
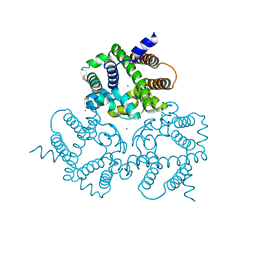 | | Crystal Structure Analysis of a Eukaryotic Membrane Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7D2Y
 
 | | complex of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, RRM2, SULFATE ION | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7D1L
 
 | | complex structure of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, Uncharacterized protein | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7EJS
 
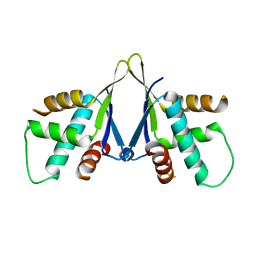 | | Structure of ERH-2 bound to PICS-1 | | Descriptor: | Enhancer of rudimentary homolog 2,Protein pid-3 | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.387 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7EJO
 
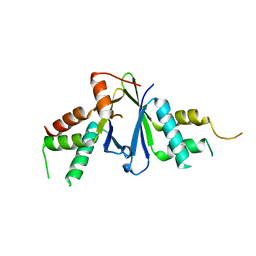 | | Structure of ERH-2 bound to TOST-1 | | Descriptor: | Enhancer of rudimentary homolog 2, Enhancer of rudimentary homolog 2,Protein tost-1 | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
5Y81
 
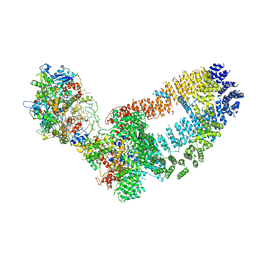 | | NuA4 TEEAA sub-complex | | Descriptor: | Actin, Actin-related protein 4, Chromatin modification-related protein EAF1, ... | | Authors: | Wang, X, Cai, G. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex
Nat Commun, 9, 2018
|
|
7FC3
 
 | | structure of NL63 receptor-binding domain complexed with horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, X.Q, Ge, J.W, Lan, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7FC5
 
 | | Crystal structure of SARS-CoV-2 RBD and horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1 | | Authors: | Wang, X.Q, Lan, J, Ge, J.W. | | Deposit date: | 2021-07-13 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
