6QUY
 
 | | NgCKK (N.Gruberi CKK) decorated 13pf taxol-GDP microtubule | | Descriptor: | CKK domain protein, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QVE
 
 | | NgCKK (Naegleria Gruberi CKK) decorated 14pf taxol-GDP microtubule | | Descriptor: | Beta1-tubulin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
1GYX
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZOIC ACID, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
4JLW
 
 | | Crystal structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Glutathione-independent formaldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Chen, S, Liao, Y.P, Wang, D.L, Wang, S, Ding, J.F, Wang, Y.M, Cai, L.J, Ran, X.Y, Zhu, H.X. | | Deposit date: | 2013-03-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa: the binary complex with the cofactor NAD+.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1GYY
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1XOG
 
 | | N9 Tern Influenza neuraminidase complexed with a 2,5-Disubstituted tetrahydrofuran-5-carboxylic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]-2,5-ANHYDRO-3,4-DIDEOXY-4-(METHOXYCARBONYL)PENTONIC ACID, Neuraminidase, ... | | Authors: | Wang, G.T, Wang, S, Gentles, R, Sowin, T, Maring, C.J, Kempf, D.J, Kati, W.M, Stoll, V, Stewart, K.D, Laver, G. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, synthesis, and structural analysis of inhibitors of influenza neuraminidase containing a 2,3-disubstituted tetrahydrofuran-5-carboxylic acid core.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XOE
 
 | | N9 Tern influenza neuraminidase complexed with (2R,4R,5R)-5-(1-Acetylamino-3-methyl-butyl-pyrrolidine-2, 4-dicarobyxylic acid 4-methyl esterdase complexed with | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]-4-(METHOXYCARBONYL)PROLINE, Neuraminidase, ... | | Authors: | Wang, G.T, Wang, S, Gentles, R, Sowin, T, Maring, C.J, Kempf, D.J, Kati, W.M, Stoll, V, Stewart, K.D, Laver, G. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and structural analysis of inhibitors of influenza neuraminidase
containing a 2,3-disubstituted tetrahydrofuran-5-carboxylic acid core.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1GYJ
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-23 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1SJU
 
 | | MINI-PROINSULIN, SINGLE CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP AND PEPTIDE BOND BETWEEN LYS B 29 AND GLY A 1, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
1SJT
 
 | | MINI-PROINSULIN, TWO CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
6S0K
 
 | | Ribosome nascent chain in complex with SecA | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Jomaa, A, Wang, S, Shan, S, Ban, N. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The molecular mechanism of cotranslational membrane protein recognition and targeting by SecA.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
1U7H
 
 | | Structure and a Proposed Mechanism for Ornithine Cyclodeaminase from Pseudomonas putida | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Alam, S, Goodman, J.L, Wang, S, Ruzicka, F.J, Frey, P.A, Wedekind, J.E. | | Deposit date: | 2004-08-03 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ornithine Cyclodeaminase: Structure, Mechanism of Action, and Implications for
the u-Crystallin Family;
Biochemistry, 43, 2004
|
|
7T7Q
 
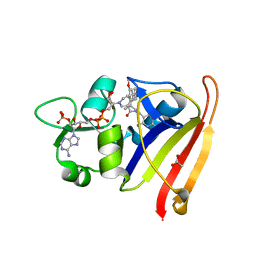 | | R-27 In Complex with S. aureus DHFR and alpha-NADPH - Remediated for comparison with tNADPH | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Reeve, S.M, Wang, S, Donald, B.R, Wright, D.L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chiral evasion and stereospecific antifolate resistance in Staphylococcus aureus.
Plos Comput.Biol., 18, 2022
|
|
1PJD
 
 | | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers | | Descriptor: | Pheromone alpha factor receptor | | Authors: | Valentine, K.G, Liu, S.-F, Marassi, F.M, Veglia, G, Nevzorov, A.A, Opella, S.J, Ding, F.-X, Wang, S.-H, Arshava, B, Becker, J.M, Naider, F. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure and Topology of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor in Phospholipid Bilayers
Biopolymers, 59, 2001
|
|
1BQJ
 
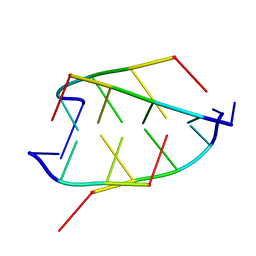 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Yang, C, Wang, S, Sutherland, C, Sinha, N, Kang, C.H. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7T7S
 
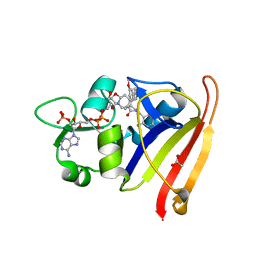 | | R-27 in Complex with S. aureus DHFR and tricyclic-NADPH (tNADPH) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Reeve, S.M, Wang, S, Donald, B.R, Wright, D.L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chiral evasion and stereospecific antifolate resistance in Staphylococcus aureus.
Plos Comput.Biol., 18, 2022
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
8FCV
 
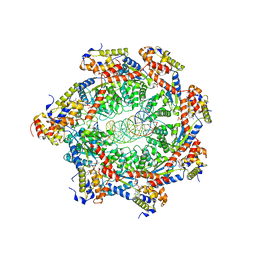 | |
4YNB
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2015-03-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4YO8
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)(hexyl)amino)methanol | | Descriptor: | Aminodeoxyfutalosine nucleosidase, ZINC ION, {[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl](hexyl)amino}methanol | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2015-03-11 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
8FCW
 
 | |
8FCU
 
 | |
8FCX
 
 | |
