7XZC
 
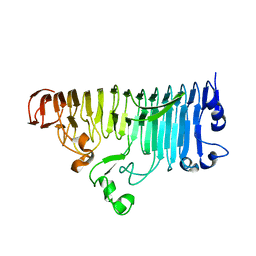 | |
4NX2
 
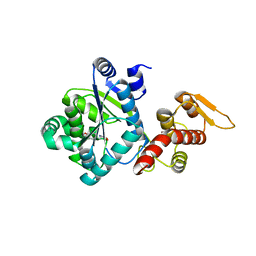 | | Crystal structure of DCYRS complexed with DCY | | Descriptor: | 3,5-dichloro-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Wang, J, Gong, W, Li, J, Gao, F, Li, H. | | Deposit date: | 2013-12-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXG
 
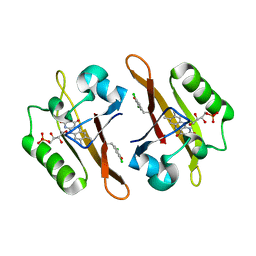 | | Crystal structure of iLOV-I486z(2LT) at pH 9.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXB
 
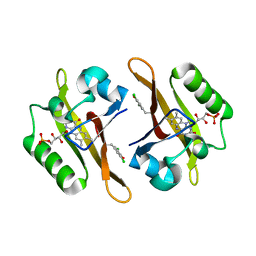 | | Crystal structure of iLOV-I486(2LT) at pH 7.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Li, J, Liu, X. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
1JCX
 
 | | Aquifex aeolicus KDO8P synthase in complex with API and Cadmium | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, {[(2,2-DIHYDROXY-ETHYL)-(2,3,4,5-TETRAHYDROXY-6-PHOSPHONOOXY-HEXYL)-AMINO]-METHYL}-PHOSPHONIC ACID | | Authors: | Wang, J, Duewel, H.S, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2001-06-11 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Aquifex aeolicus KDO8P synthase in complex with R5P and PEP, and with a bisubstrate inhibitor: role of active site water in catalysis.
Biochemistry, 40, 2001
|
|
4NXE
 
 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
1JCY
 
 | | Aquifex aeolicus KDO8P synthase in complex with R5P, PEP and Cadmium | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Duewel, H.S, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2001-06-11 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Aquifex aeolicus KDO8P synthase in complex with R5P and PEP, and with a bisubstrate inhibitor: role of active site water in catalysis.
Biochemistry, 40, 2001
|
|
1ZA1
 
 | | Structure of wild-type E. coli Aspartate Transcarbamoylase in the presence of CTP at 2.20 A resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, J, Stieglitz, K.A, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5UY8
 
 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | 5-[(5S)-5-ethyl-5-methyl-6-oxo-1,4,5,6-tetrahydropyridin-3-yl]-N-(6-fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)thiophene-2-sulfonamide, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, ... | | Authors: | Wang, J, Wang, Y, Fales, K.R, Atwell, S, Clawson, D. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
4L44
 
 | | Crystal structures of human p70S6K1-T389A (form II) | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein, SULFATE ION, ... | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
4L42
 
 | | Crystal structures of human p70S6K1-PIF | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein, SULFATE ION, ... | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
4L46
 
 | | Crystal structures of human p70S6K1-WT | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein, SULFATE ION, ... | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
4L45
 
 | | Crystal structures of human p70S6K1-T389E | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
4L43
 
 | | Crystal structures of human p70S6K1-T389A (form I) | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
1JOR
 
 | | Ensemble structures for unligated Staphylococcal nuclease-H124L | | Descriptor: | staphylococcal nuclease | | Authors: | Wang, J, Truckses, D.M, Abildgaard, F, Dzakula, Z, Zolnai, Z, Markley, J.L. | | Deposit date: | 2001-07-30 | | Release date: | 2001-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of staphylococcal nuclease from multidimensional, multinuclear NMR: nuclease-H124L and its ternary complex with Ca2+ and thymidine-3',5'-bisphosphate.
J.Biomol.NMR, 10, 1997
|
|
1XK3
 
 | | NADPH- and Ascorbate-Supported Heme Oxygenase Reactions are Distinct. Regiospecificity of Heme Cleavage by the R183E Mutant | | Descriptor: | Heme oxygenase 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Lad, L, Poulos, T.L, Ortiz de montellano, P.R. | | Deposit date: | 2004-09-26 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Regiospecificity determinants of human heme oxygenase: differential NADPH- and ascorbate-dependent heme cleavage by the R183E mutant.
J.Biol.Chem., 280, 2005
|
|
1XK2
 
 | | NADPH- and Ascorbate-Supported Heme Oxygenase Reactions are Distinct. Regiospecificity of Heme Cleavage by the R183E Mutant | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Lad, L, Poulos, T.L, Ortiz de montellano, P.R. | | Deposit date: | 2004-09-26 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regiospecificity determinants of human heme oxygenase: differential NADPH- and ascorbate-dependent heme cleavage by the R183E mutant.
J.Biol.Chem., 280, 2005
|
|
4L3L
 
 | | Crystal structures of human p70S6K1 kinase domain (Zinc anomalous) | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein, ZINC ION | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
4L3J
 
 | | Crystal structures of human p70S6K1 kinase domain | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein, ZINC ION | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
1ZA2
 
 | | Structure of wild-type E. coli Aspartate Transcarbamoylase in the presence of CTP, carbamoyl phosphate at 2.50 A resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, J, Stieglitz, K.A, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZET
 
 | |
1JOO
 
 | | Averaged structure for unligated Staphylococcal nuclease-H124L | | Descriptor: | staphylococcal nuclease | | Authors: | Wang, J, Truckses, D.M, Abildgaard, F, Dzakula, Z, Zolnai, Z, Markley, J.L. | | Deposit date: | 2001-07-30 | | Release date: | 2001-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of staphylococcal nuclease from multidimensional, multinuclear NMR: nuclease-H124L and its ternary complex with Ca2+ and thymidine-3',5'-bisphosphate.
J.Biomol.NMR, 10, 1997
|
|
1K0J
 
 | | Pseudomonas aeruginosa phbh R220Q in complex with NADPH and free of p-OHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, P-HYDROXYBENZOATE HYDROXYLASE, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
