7DFV
 
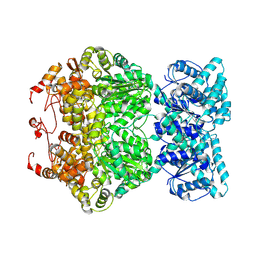 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7CR7
 
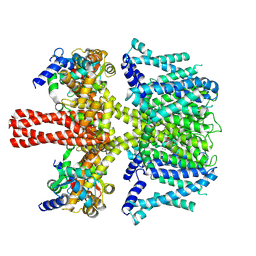 | | human KCNQ2-CaM in complex with retigabine | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR0
 
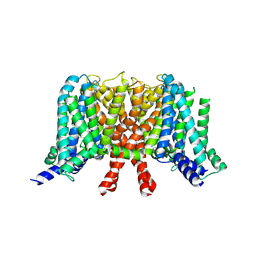 | | human KCNQ2 in apo state | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR3
 
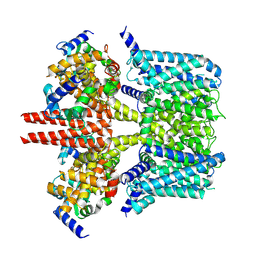 | | human KCNQ2-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
2KJG
 
 | |
1G6M
 
 | |
2LA9
 
 | | NMR structure of Pseudouridine_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*(PSU)P*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
7CRB
 
 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | Descriptor: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7CR4
 
 | | human KCNQ2-CaM in complex with ztz240 | | Descriptor: | Calmodulin-3, N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR1
 
 | | human KCNQ2 in complex with ztz240 | | Descriptor: | N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
2LEO
 
 | |
2KHS
 
 | | Solution structure of SNase121:SNase(111-143) complex | | Descriptor: | Nuclease, Thermonuclease | | Authors: | Geng, Y, Feng, Y, Xie, T, Shan, L, Wang, J. | | Deposit date: | 2009-04-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The native-like interactions between SNase121 and SNase(111-143) fragments induce the recovery of their native-like structures and the ability to degrade DNA.
Biochemistry, 48, 2009
|
|
2PZY
 
 | | Structure of MK2 Complexed with Compound 76 | | Descriptor: | (4R)-N-[4-({[2-(DIMETHYLAMINO)ETHYL]AMINO}CARBONYL)-1,3-THIAZOL-2-YL]-4-METHYL-1-OXO-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLINE-6-CARBOXAMIDE, MAP kinase-activated protein kinase 2, STAUROSPORINE | | Authors: | White, A, Wu, J.P, Wang, J, Abeywardane, A, Andersen, D, Emmanuel, M, Gautschi, E, Goldberg, D.R, Kashem, M.A, Lukas, S, Mao, W, Martin, L, Morwick, T, Moss, N, Pargellis, C, Patel, U.R, Patnaude, L, Peet, G.W, Skow, D, Snow, R.J, Ward, Y, Werneburg, B. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The discovery of carboline analogs as potent MAPKAP-K2 inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7C4U
 
 | | MicroED structure of orthorhombic Vancomycin at 1.2 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7C4V
 
 | | MicroED structure of anorthic Vancomycin at 1.05 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7DZH
 
 | | intermediate of FABP with a delay time of 100 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZE
 
 | | Fabp ground state captured by XFELs | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZF
 
 | | Intermediate of FABP with a delay time of 10 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZG
 
 | | Intermediate of FABP with a delay time of 30 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7EA9
 
 | | Crystal Structure of human lysyl-tRNA synthetase Y145H mutant | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, GLYCEROL, Lysine--tRNA ligase | | Authors: | Wu, S, Hei, Z, Zheng, L, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analyses of a human lysyl-tRNA synthetase mutant associated with autosomal recessive nonsyndromic hearing impairment.
Biochem.Biophys.Res.Commun., 554, 2021
|
|
7E2M
 
 | | Crystal structure of the RWD domain of human GCN2 - 2 | | Descriptor: | eIF-2-alpha kinase GCN2 | | Authors: | Hei, Z, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures reveal a novel dimer of the RWD domain of human general control nonderepressible 2.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
7E2K
 
 | | Crystal structure of the RWD domain of human GCN2 - 1 | | Descriptor: | eIF-2-alpha kinase GCN2 | | Authors: | Hei, Z, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Crystal structures reveal a novel dimer of the RWD domain of human general control nonderepressible 2.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
7CUN
 
 | | The structure of human Integrator-PP2A complex | | Descriptor: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | Authors: | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
7DZI
 
 | | intermediate of FABP with a delay time of 300 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
