4BEV
 
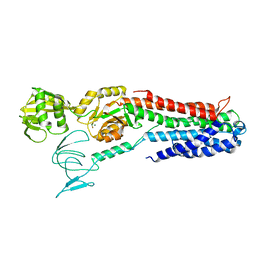 | | ATPase crystal structure with bound phosphate analogue | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, TRIFLUOROMAGNESATE | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Gourdon, P, Pedersen, B.P, Morth, P, Wang, J, Nissen, P. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | ATPase Crystal Structure with Bound Phosphate Analogue
To be Published
|
|
5NZ3
 
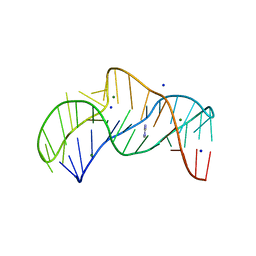 | | The structure of the thermobifida fusca guanidine III riboswitch with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
3FWE
 
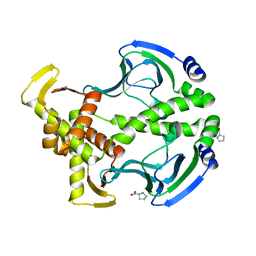 | | Crystal Structure of the Apo D138L CAP mutant | | Descriptor: | Catabolite gene activator, PROLINE | | Authors: | Sharma, H, Wang, J, Kong, J, Yu, S, Steitz, T. | | Deposit date: | 2009-01-17 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5NY8
 
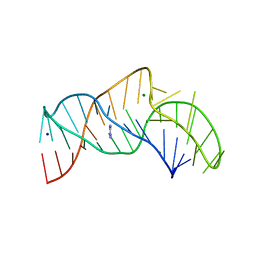 | | The structure of the thermobifida fusca guanidine III riboswitch with aminoguanidine | | Descriptor: | AMINOGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NZ6
 
 | |
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
5NZD
 
 | | The structure of the thermobifida fusca guanidine III riboswitch in space group P212121. | | Descriptor: | ACETATE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-13 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5NWQ
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with guanidine. | | Descriptor: | GUANIDINE, Guanidine III riboswitch, MAGNESIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-08 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
6IFU
 
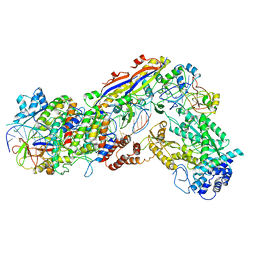 | | Cryo-EM structure of type III-A Csm-CTR2-dsDNA complex | | Descriptor: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
3NFP
 
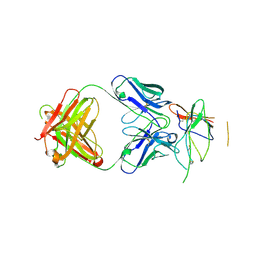 | | Crystal structure of the Fab fragment of therapeutic antibody daclizumab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Interleukin-2 receptor subunit alpha, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISI
 
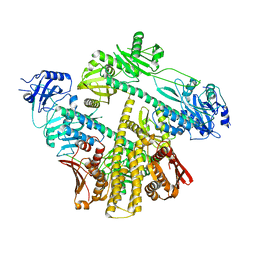 | | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | | Descriptor: | Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
5MGW
 
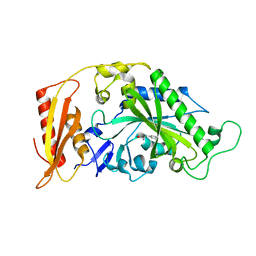 | | Kinetic and Structural Changes in HsmtPheRS, Induced by Pathogenic Mutations in Human FARS2 | | Descriptor: | PHENYLALANINE, Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
5O62
 
 | |
5O69
 
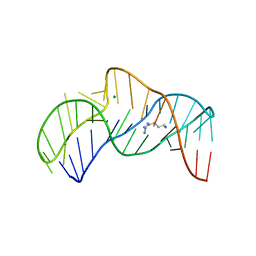 | | The structure of the thermobifida fusca guanidine III riboswitch with agmatine. | | Descriptor: | AGMATINE, MAGNESIUM ION, RNA (37-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
4AW2
 
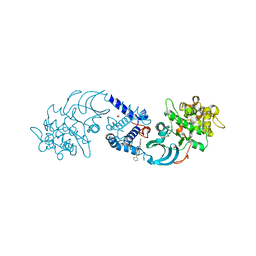 | | Crystal structure of CDC42 binding protein kinase alpha (MRCK alpha) | | Descriptor: | 1,2-ETHANEDIOL, 5,11-dimethyl-1-oxo-2,6-dihydro-1h-pyrido[4,3-b]carbazol-9-yl benzoate, SERINE/THREONINE-PROTEIN KINASE MRCK ALPHA | | Authors: | Elkins, J.M, Muniz, J.R.C, Tan, I, Leung, T, Lafanechere, L, Prudent, R, Abdul Azeez, K, Szklarz, M, Phillips, C, Wang, J, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cdc42 Binding Protein Kinase Alpha (Mrck Alpha)
To be Published
|
|
4DTO
 
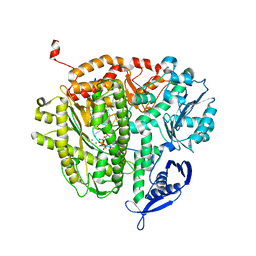 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite an Abasic Site and ddA/dT as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DTJ
 
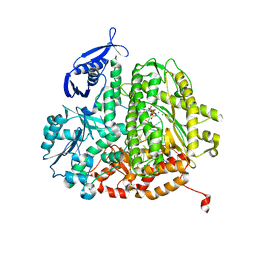 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite an Abasic Site and ddT/dA as the Penultimate Base-pair | | Descriptor: | CALCIUM ION, DNA polymerase, DNA primer, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DU3
 
 | | RB69 DNA Polymerase Ternary Complex with dDTP Opposite dT with 3-Deaza-adenine at the N-1 Position of Template Strand | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA.
Biochemistry, 51, 2012
|
|
4DTR
 
 | | RB69 DNA Polymerase Ternary Complex with dATP Opposite an Abasic Site and ddC/dG as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DWQ
 
 | | RNA ligase RtcB-GMP/Mn(2+) complex | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MALONATE ION, ... | | Authors: | Okada, C, Xia, S, Englert, M, Yao, M, Soll, D, Wang, J. | | Deposit date: | 2012-02-26 | | Release date: | 2012-09-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DTM
 
 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite an Abasic Site and ddG/dC as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
4DU4
 
 | | RB69 DNA Polymerase Ternary Complex with dATP Opposite dT with 3-Deaza-adenine at the N-3 Position of Primer Strand | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA.
Biochemistry, 51, 2012
|
|
4DTP
 
 | | RB69 DNA Polymerase Ternary Complex with dGTP Opposite an Abasic Site and ddA/dT as the Penultimate Base-pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of Partial Charge Interactions and Base Stacking to the Efficiency of Primer Extension at and beyond Abasic Sites in DNA.
Biochemistry, 51, 2012
|
|
