6IFL
 
 | | Cryo-EM structure of type III-A Csm-NTR complex | | 分子名称: | NTR, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
7EOD
 
 | | MITF HLHLZ Delta AKE | | 分子名称: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | 著者 | Li, P, Liu, Z, Fang, P, Wang, J. | | 登録日 | 2021-04-22 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
4M9Y
 
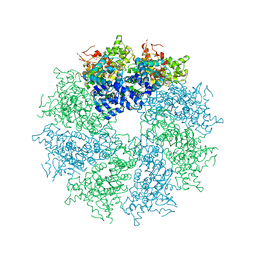 | | Crystal structure of CED-4 bound CED-3 fragment | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | 著者 | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | 登録日 | 2013-08-15 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
4MSR
 
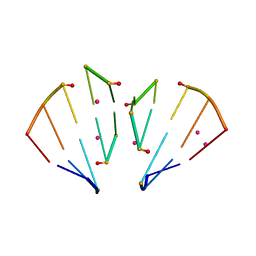 | | RNA 10mer duplex with six 2'-5'-linkages | | 分子名称: | RNA 10mer duplex with six 2'-5'-linkages, STRONTIUM ION | | 著者 | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | 登録日 | 2013-09-18 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6IIL
 
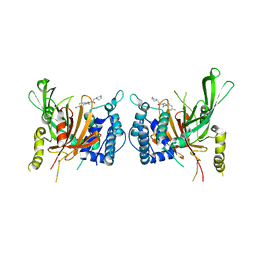 | | USP14 catalytic domain bind to IU1-47 | | 分子名称: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(piperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | 著者 | Mei, Z.Q, Wang, Y.W, Wang, F, Wang, J.W, He, W, Ding, S, Li, J.W. | | 登録日 | 2018-10-07 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
1ECL
 
 | |
1EFG
 
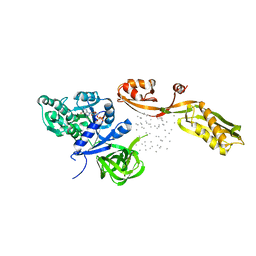 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR G COMPLEXED WITH GDP, AT 2.7 ANGSTROMS RESOLUTION | | 分子名称: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | 登録日 | 1994-10-17 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
5TW5
 
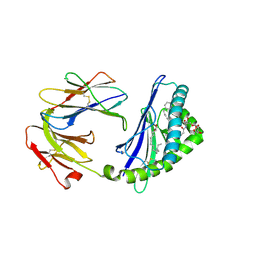 | | Structure of mouse CD1d with bound glycosphingolipid JJ112 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | 著者 | Zajonc, D.M, Wang, J. | | 登録日 | 2016-11-11 | | 公開日 | 2017-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Galactosylsphingamides: new alpha-GalCer analogues to probe the F'-pocket of CD1d.
Sci Rep, 7, 2017
|
|
7TQL
 
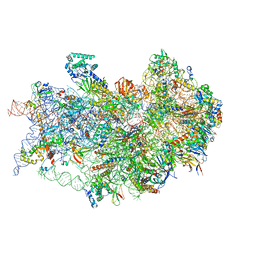 | | CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B. | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Lapointe, C.P, Grosely, R, Sokabe, M, Alvarado, C, Wang, J, Montabana, E, Villa, N, Shin, B, Dever, T, Fraser, C, Fernandez, I.S, Puglisi, J.D. | | 登録日 | 2022-01-26 | | 公開日 | 2022-04-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining.
Nature, 607, 2022
|
|
6J6K
 
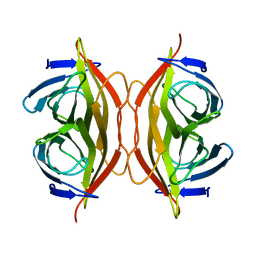 | | Apo-state streptavidin | | 分子名称: | Streptavidin | | 著者 | Fan, X, Wang, J, Lei, J.L, Wang, H.W. | | 登録日 | 2019-01-15 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.
Nat Commun, 10, 2019
|
|
6J6J
 
 | | Biotin-bound streptavidin | | 分子名称: | BIOTIN, Streptavidin | | 著者 | Fan, X, Wang, J, Lei, J.L, Wang, H.W. | | 登録日 | 2019-01-15 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.
Nat Commun, 10, 2019
|
|
6JEA
 
 | | crystal structure of a beta-N-acetylhexosaminidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ZINC ION | | 著者 | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | 登録日 | 2019-02-04 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.275 Å) | | 主引用文献 | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7EQH
 
 | |
7EOZ
 
 | |
7TOB
 
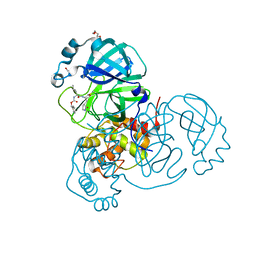 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Sacco, M.D, Wang, J, Chen, Y. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
8XPN
 
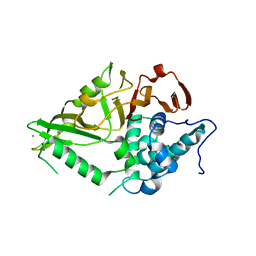 | | The Crystal Structure of USP8 from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ubiquitin carboxyl-terminal hydrolase 8, ... | | 著者 | Wang, F, Cheng, W, Yuan, Z, Lin, D, Wang, J. | | 登録日 | 2024-01-04 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of USP8 from Biortus.
To Be Published
|
|
8YGZ
 
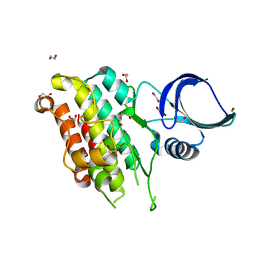 | | The Crystal Structure of TGF beta R2 kinase domain from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, TGF-beta receptor type-2 | | 著者 | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | 登録日 | 2024-02-27 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of TGF beta R2 kinase domain from Biortus.
To Be Published
|
|
2PH7
 
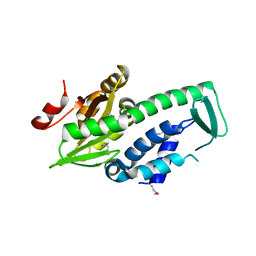 | | Crystal structure of AF2093 from Archaeoglobus fulgidus | | 分子名称: | Uncharacterized protein AF_2093 | | 著者 | Chang, J.C, Yang, H, Hwang, J, Zhu, J, Chen, L, Fu, Z.-Q, Xu, H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2007-04-10 | | 公開日 | 2007-05-08 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of AF2093 from Archaeoglobus fulgidus.
To be Published
|
|
8XCG
 
 | |
4CDQ
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | 分子名称: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | 著者 | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-05 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CEW
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | 分子名称: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-12 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | 分子名称: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-12 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDX
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | 分子名称: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-07 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDW
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | 分子名称: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-06 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDU
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | 分子名称: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | 著者 | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2013-11-06 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
