3A7F
 
 | | Human MST3 kinase | | Descriptor: | Serine/threonine kinase 24 (STE20 homolog, yeast) | | Authors: | Ko, T.P, Jeng, W.Y, Liu, C.I, Lai, M.D, Wang, A.H.J. | | Deposit date: | 2009-09-26 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of human MST3 kinase in complex with adenine, ADP and Mn2+.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A7H
 
 | | Human MST3 kinase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Serine/threonine kinase 24 (STE20 homolog, yeast) | | Authors: | Ko, T.P, Jeng, W.Y, Liu, C.I, Lai, M.D, Wang, A.H.J. | | Deposit date: | 2009-09-26 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of human MST3 kinase in complex with adenine, ADP and Mn2+.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5ITJ
 
 | | The structure of histone-like protein | | Descriptor: | AbrB family transcriptional regulator, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
1KUK
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEKW. | | Descriptor: | CADMIUM ION, EKW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
5ITM
 
 | | The structure of truncated histone-like protein | | Descriptor: | AbrB family transcriptional regulator | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
1KUG
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with its endogenous inhibitor pENW | | Descriptor: | CADMIUM ION, ENW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUF
 
 | | High-resolution Crystal Structure of a Snake Venom Metalloproteinase from Taiwan Habu | | Descriptor: | CADMIUM ION, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Yuann, J.M, Wang, A.H.J. | | Deposit date: | 2002-01-21 | | Release date: | 2002-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The 1.35 A structure of cadmium-substituted TM-3, a snake-venom metalloproteinase from Taiwan habu: elucidation of a TNFalpha-converting enzyme-like active-site structure with a distorted octahedral geometry of cadmium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KUI
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEQW. | | Descriptor: | CADMIUM ION, EQW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
2ZB7
 
 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADPH and nicotinamide | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, Prostaglandin reductase 2 | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
3A7G
 
 | | Human MST3 kinase | | Descriptor: | Serine/threonine kinase 24 (STE20 homolog, yeast) | | Authors: | Ko, T.P, Jeng, W.Y, Liu, C.I, Lai, M.D, Wang, A.H.J. | | Deposit date: | 2009-09-26 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of human MST3 kinase in complex with adenine, ADP and Mn2+.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1JP3
 
 | | Structure of E.coli undecaprenyl pyrophosphate synthase | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, undecaprenyl pyrophosphate synthase | | Authors: | Ko, T.P, Chen, Y.K, Robinson, H, Tsai, P.C, Gao, Y.G, Chen, A.P.C, Wang, A.H.J, Liang, P.H. | | Deposit date: | 2001-07-31 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis.
J.Biol.Chem., 276, 2001
|
|
2QN5
 
 | | Crystal Structure and Functional Study of the Bowman-Birk Inhibitor from Rice Bran in Complex with Bovine Trypsin | | Descriptor: | Bowman-Birk type bran trypsin inhibitor, Cationic trypsin | | Authors: | Li, H.T, Lin, Y.H, Guan, H.H, Hsieh, Y.C, Wang, A.H.J, Chen, C.J. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure and Functional Study of the Bowman-Birk Inhibitor from Rice Bran in Complex with Bovine Trypsin
To be Published
|
|
3VJE
 
 | | Crystal structure of the Y248A mutant of C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus in complex with zaragozic acid A | | Descriptor: | Dehydrosqualene synthase, Zaragozic acid A | | Authors: | Liu, C.I, Jeng, W.Y, Chang, W.J, Wang, A.H.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
3VJD
 
 | | Crystal structure of the Y248A mutant of C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus | | Descriptor: | Dehydrosqualene synthase, L(+)-TARTARIC ACID | | Authors: | Liu, C.I, Jeng, W.Y, Chang, W.J, Wang, A.H.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
3W1O
 
 | | Neisseria DNA mimic protein DMP12 | | Descriptor: | DNA mimic protein DMP12, MAGNESIUM ION | | Authors: | Wang, H.C, Ko, T.P, Wu, M.L, Wang, A.H.J. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein
Nucleic Acids Res., 41, 2013
|
|
3WUR
 
 | | Structure of DMP19 Complex with 18-crown-6 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, L(+)-TARTARIC ACID, ... | | Authors: | Lee, C.C, Wang, H.C, Wang, A.H.J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1UDV
 
 | | Crystal structure of the hyperthermophilic archaeal dna-binding protein Sso10b2 at 1.85 A | | Descriptor: | DNA binding protein SSO10b, ZINC ION | | Authors: | Chou, C.-C, Lin, T.-W, Chen, C.-Y, Wang, A.H.J. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms
J.BACTERIOL., 185, 2003
|
|
3VOP
 
 | | Structure of Vaccinia virus A27 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein A27 | | Authors: | Chang, T.H, Ko, T.P, Hsieh, F.L, Wang, A.H.J. | | Deposit date: | 2012-01-31 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of vaccinia viral A27 protein reveals a novel structure critical for its function and complex formation with A26 protein.
Plos Pathog., 9, 2013
|
|
3WX4
 
 | |
3WDG
 
 | | Staphylococcus aureus UDG / UGI complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
3WDF
 
 | | Staphylococcus aureus UDG | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
3WH0
 
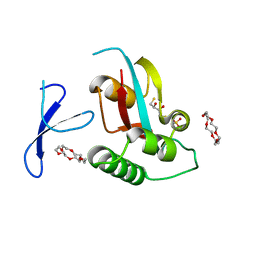 | | Structure of Pin1 Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Lee, C.C, Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WHM
 
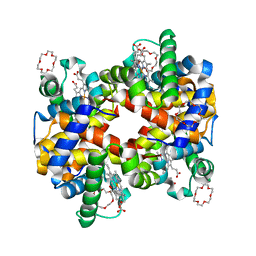 | | Structure of Hemoglobin Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Lee, C.C, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7C8T
 
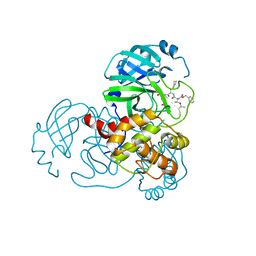 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0205221 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
7C8R
 
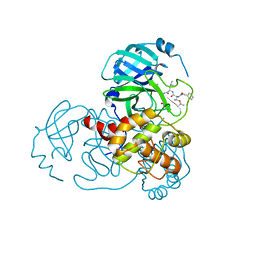 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0203770 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
