2R8Q
 
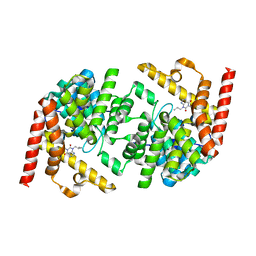 | | Structure of LmjPDEB1 in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, Class I phosphodiesterase PDEB1, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Geng, J, Kunz, S, Seebeck, T, Ke, H. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Leishmania major phosphodiesterase LmjPDEB1 and insight into the design of the parasite-selective inhibitors.
Mol.Microbiol., 66, 2007
|
|
2CW4
 
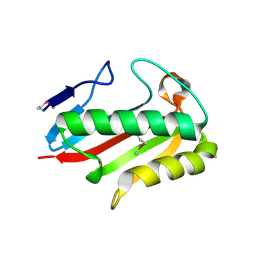 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-16 | | Release date: | 2005-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
2CVL
 
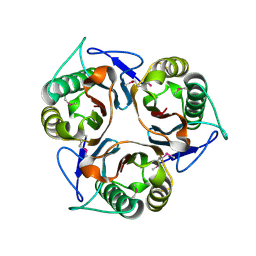 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | protein translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
3O47
 
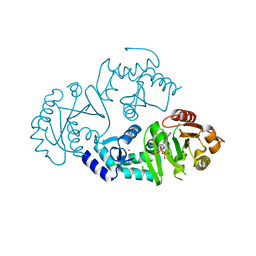 | | Crystal structure of ARFGAP1-ARF1 fusion protein | | Descriptor: | ADP-ribosylation factor GTPase-activating protein 1, ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Tong, Y, Nedyalkova, L, Tempel, W, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ARFGAP1-ARF1 fusion protein
to be published
|
|
2YV5
 
 | | Crystal structure of Yjeq from Aquifex aeolicus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, YjeQ protein, ... | | Authors: | Wang, H, Kaminishi, T, Hanawa-Suetsugu, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of YjeQ from Aquifex aeolicus
To be Published
|
|
2YVR
 
 | | Crystal structure of MS1043 | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of MS1043
To be Published
|
|
4MM4
 
 | |
4MM8
 
 | |
4MMF
 
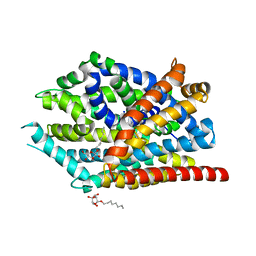 | | Crystal structure of LeuBAT (delta5 mutant) in complex with mazindol | | Descriptor: | (5R)-5-(4-chlorophenyl)-2,5-dihydro-3H-imidazo[2,1-a]isoindol-5-ol, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MM9
 
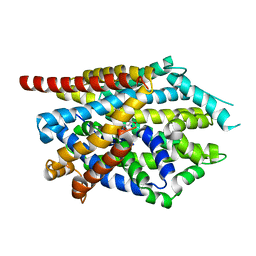 | |
2Z0I
 
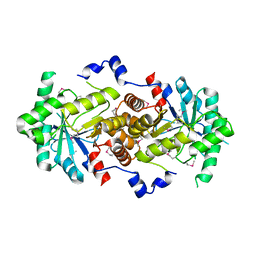 | | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculus | | Descriptor: | Delta-aminolevulinic acid dehydratase | | Authors: | Wang, H, Xie, Y, Kawazoe, M, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculus
To be Published
|
|
3PFN
 
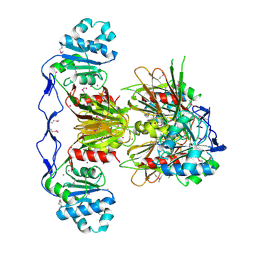 | | Crystal Structure of human NAD kinase | | Descriptor: | NAD kinase, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Tempel, W, Wernimont, A.K, Tong, Y, Guan, X, Shen, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of human NAD kinase
to be published
|
|
3Q3J
 
 | | Crystal structure of plexin A2 RBD in complex with Rnd1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-A2, ... | | Authors: | Wang, H, Tempel, W, Tong, Y, Guan, X, Shen, L, Buren, L, Zhang, N, Wernimont, A.K, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-21 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Crystal structure of plexin A2 RBD in complex with Rnd1
to be published
|
|
6LHE
 
 | | Crystal Structure of Gold-bound NDM-1 | | Descriptor: | GOLD ION, Metallo-beta-lactamase type 2, SULFATE ION | | Authors: | Wang, H, Sun, H, Wang, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
5J07
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P1/2 | | Descriptor: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | Deposit date: | 2016-03-27 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5J0C
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P2/3 | | Descriptor: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn],OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
4MMD
 
 | |
2RIB
 
 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with L-glycero-D-manno-heptose | | Descriptor: | CALCIUM ION, L-glycero-alpha-D-manno-heptopyranose, Pulmonary surfactant-associated protein D | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
2ZUL
 
 | | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) in complex with cofactor S-adenosyl-L-Methionine | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Wang, H, Kawazoe, M, Tatsuguchi, A, Naoe, C, Takemoto, C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-21 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) in complex with cofactor S-adenosyl-L-Methionine
To be Published
|
|
5J0G
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P7/8 | | Descriptor: | OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5J0F
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P4/5 | | Descriptor: | GLYCEROL, Superoxide dismutase [Cu-Zn],OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
4MM5
 
 | | Crystal structure of LeuBAT (delta13 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MMB
 
 | | Crystal structure of LeuBAT (delta6 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MMA
 
 | |
2RID
 
 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with Allyl 7-O-carbamoyl-L-glycero-D-manno-heptopyranoside | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, prop-2-en-1-yl 7-O-carbamoyl-L-glycero-alpha-D-manno-heptopyranoside | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
