5WJT
 
 | |
5XSI
 
 | | The catalytic domain of GdpP | | Descriptor: | MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5WK5
 
 | |
5WJX
 
 | | Cryo-EM structure of B. subtilis flagellar filaments S17P | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJW
 
 | | Cryo-EM structure of B. subtilis flagellar filaments H84R | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJV
 
 | |
5WJZ
 
 | | Cryo-EM structure of B. subtilis flagellar filaments E115G | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5XSN
 
 | | The catalytic domain of GdpP with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XBW
 
 | | The structure of BrlR | | Descriptor: | Probable transcriptional regulator | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
5XBI
 
 | |
5XBT
 
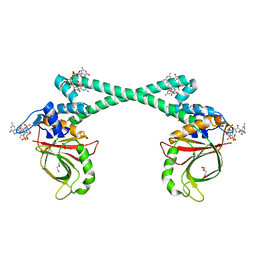 | | The structure of BrlR bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
2I46
 
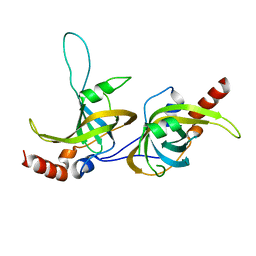 | | Crystal structure of human TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog | | Authors: | Wang, F, Podell, E.R, Zaug, A.J, Yang, Y.T, Baciu, P, Else, T, Hammer, G.D, Cech, T.R, Lei, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The POT1-TPP1 telomere complex is a telomerase processivity factor.
Nature, 445, 2007
|
|
2HVC
 
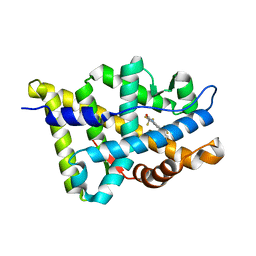 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | Descriptor: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | Authors: | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | Deposit date: | 2006-07-28 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2H9I
 
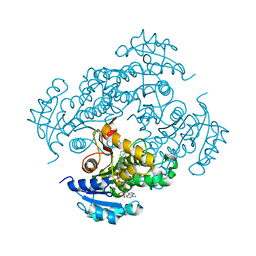 | | Mycobacterium tuberculosis InhA bound with ETH-NAD adduct | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], {(2R,3S,4R,5R)-5-[(4S)-3-(AMINOCARBONYL)-4-(2-ETHYLISONICOTINOYL)PYRIDIN-1(4H)-YL]-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL}METHYL [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2006-06-09 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of thioamide drug action against tuberculosis and leprosy.
J.Exp.Med., 204, 2007
|
|
2LQI
 
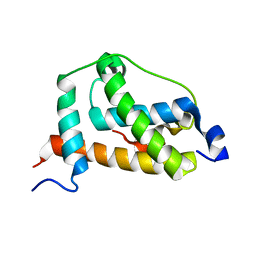 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LQH
 
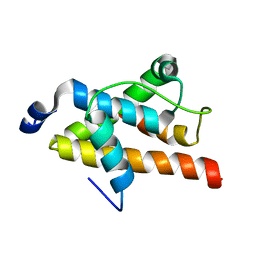 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7ESE
 
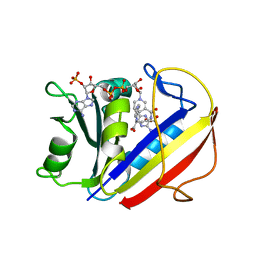 | | The Crystal Structure of human DHFR from Biortus | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, F, Cheng, W, Xu, C, Qi, J, Bao, X, Miao, Q. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of human DHFR from Biortus
To Be Published
|
|
6XG6
 
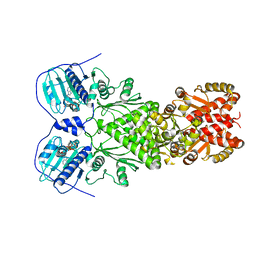 | | Full-length human mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Heat shock protein 75 kDa, ... | | Authors: | Liu, Y.X, Wang, F, Agard, D.A. | | Deposit date: | 2020-06-17 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | General and robust covalently linked graphene oxide affinity grids for high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DY7
 
 | | WDR5 in complex with a WIN site inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N-[3-(2,4-dichlorophenoxy)propyl]-1H-imidazol-2-amine, SULFATE ION, ... | | Authors: | Phan, J, Wang, F, Fesik, S.W. | | Deposit date: | 2018-07-01 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity.
Cell Rep, 26, 2019
|
|
6DYA
 
 | | WDR5 in complex with a WIN site inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3,5-dichlorophenyl)methyl]-3-[(1H-imidazol-1-yl)methyl]benzamide, SULFATE ION, ... | | Authors: | Phan, J, Wang, F, Fesik, S.W. | | Deposit date: | 2018-07-01 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity.
Cell Rep, 26, 2019
|
|
3MAQ
 
 | | Crystal structure of E.coli Pol II-normal DNA-dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*TP*GP*CP*CP*TP*AP*GP*CP*GP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*AP*CP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II
Cell(Cambridge,Mass.), 139, 2009
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | Descriptor: | GLYCEROL, Putative aminotransferase | | Authors: | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
7SLZ
 
 | | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH BPF023596 | | Descriptor: | Glucose-induced degradation protein 4 homolog, N-[(1s,4s)-4-(1H-benzimidazol-2-yl)cyclohexyl]-N~2~-[(1H-indol-2-yl)methyl]glycinamide | | Authors: | Song, X, Dong, A, Calabrese, M, Wang, F, Owen, D, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH BPF023596
To Be Published
|
|
5W5F
 
 | | Cryo-EM structure of the T4 tail tube | | Descriptor: | Tail tube protein gp19 | | Authors: | Zheng, W, Wang, F, Taylor, N.M, Guerrero-Ferreira, R.C, Leiman, P.G, Egelman, E.H. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit.
Structure, 25, 2017
|
|
7DLV
 
 | | shrimp dUTPase in complex with Stl | | Descriptor: | CALCIUM ION, Orf20, SULFATE ION, ... | | Authors: | Ma, Q, Wang, F. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of staphylococcal Stl inhibition on a eukaryotic dUTPase.
Int.J.Biol.Macromol., 184, 2021
|
|
