1CRB
 
 | |
4OLR
 
 | | [Leu-5]-Enkephalin mutant - YVVFV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, [Leu-5]-Enkephalin mutant - YVVFV | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
4ONK
 
 | | [Leu-5]-Enkephalin mutant - YVVFL | | Descriptor: | [Leu-5]-Enkephalin mutant - YVVFL | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
3T8V
 
 | | A bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases | | Descriptor: | M1 family aminopeptidase, MAGNESIUM ION, N-[(2-{2-[(N-{(2S,3R)-3-amino-4-[4-(benzyloxy)phenyl]-2-hydroxybutanoyl}-L-alanyl)amino]ethoxy}ethoxy)acetyl]-4-benzoyl-L-phenylalanyl-N~6~-hex-5-ynoyllysinamide, ... | | Authors: | McGowan, S, Klemba, M, Greebaum, D.C. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4K5P
 
 | |
4BVK
 
 | |
4K5M
 
 | |
4BVL
 
 | |
4K5L
 
 | |
4K5O
 
 | |
3Q44
 
 | |
4BRS
 
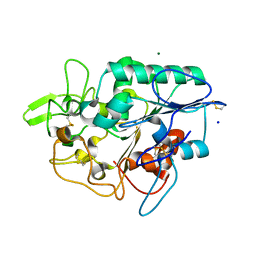 | | Structure of wild type PhaZ7 PHB depolymerase | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHB DEPOLYMERASE PHAZ7, ... | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-05 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
3Q43
 
 | |
2MBB
 
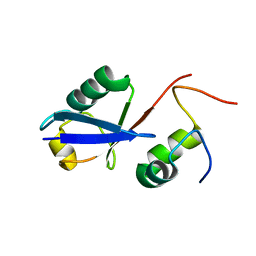 | | Solution Structure of the human Polymerase iota UBM1-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G/DNA polymerase iota fusion protein, Polyubiquitin-B | | Authors: | Wang, S, Zhou, P. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparsely-sampled, high-resolution 4-D omit spectra for detection and assignment of intermolecular NOEs of protein complexes.
J.Biomol.Nmr, 59, 2014
|
|
2OMF
 
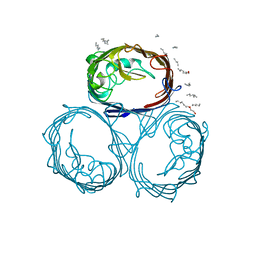 | | OMPF PORIN | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Cowan, S.W. | | Deposit date: | 1995-02-28 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Refined Structure of Ompf Porin from E.Coli at 2.4 Angstroms Resolution
To be Published
|
|
2MUR
 
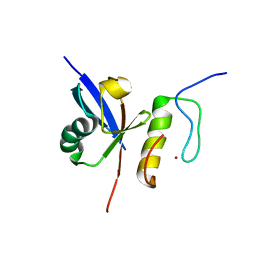 | |
4K5N
 
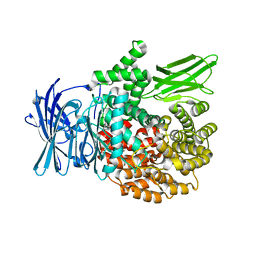 | |
4BVJ
 
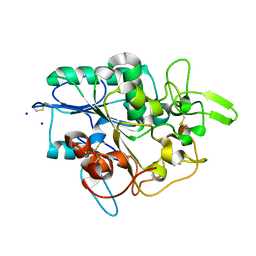 | | Structure of Y105A mutant of PhaZ7 PHB depolymerase | | Descriptor: | PHB DEPOLYMERASE PHAZ7, SODIUM ION | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
1IYB
 
 | |
3W4Y
 
 | |
1OPF
 
 | |
1VD1
 
 | |
1VCZ
 
 | |
1VD3
 
 | | Ribonuclease NT in complex with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, RNase NGR3 | | Authors: | Kawano, S, Kakuta, Y, Kimura, M. | | Deposit date: | 2004-03-18 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Nicotiana glutinosa Ribonuclease NT in Complex with Nucleotide Monophosphates
to be published
|
|
2MM6
 
 | |
