8UPR
 
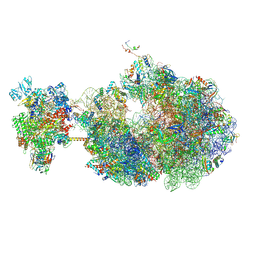 | | Escherichia coli transcription-translation coupled complex class A (TTC-A) containing RfaH bound to ops signal, mRNA with a 21 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UQM
 
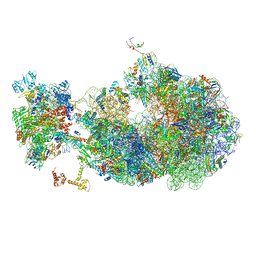 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, NusA, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
5ZQZ
 
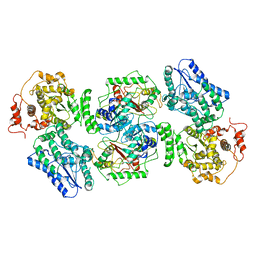 | | Structure of human mitochondrial trifunctional protein, tetramer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-20 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8H0B
 
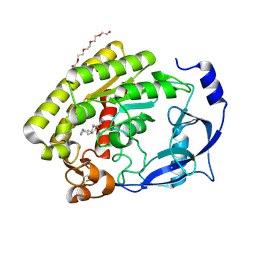 | |
8H0A
 
 | |
5ZVB
 
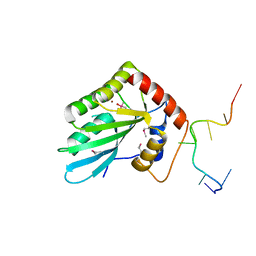 | | APOBEC3F Chimeric Catalytic Domain in Complex with DNA(dT9) | | Descriptor: | APEBEC3F/ssDNA-T9, CACODYLATE ION, DNA (5'-D(*AP*TP*TP*TP*TP*CP*AP*AP*T)-3'), ... | | Authors: | Cheng, C, Zhang, T.L, Wang, C.X, Lan, W.X, Ding, J.P, Cao, C.Y. | | Deposit date: | 2018-05-09 | | Release date: | 2018-11-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Human APOBEC3F Chimeric Catalytic Domain in Complex with DNA
Chin.J.Chem., 36, 2018
|
|
8H09
 
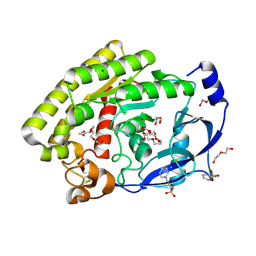 | |
8H0C
 
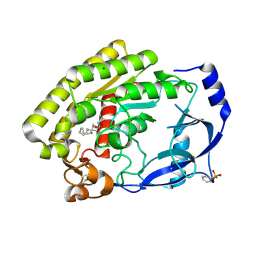 | |
8H0D
 
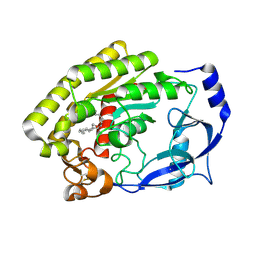 | | Structure of the thermolabile hemolysin from Vibrio alginolyticus (in complex with docosahexaenoic acid) | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, MAGNESIUM ION, ... | | Authors: | Ma, Q, Wang, C. | | Deposit date: | 2022-09-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic site flexibility facilitates the substrate and catalytic promiscuity of Vibrio dual lipase/transferase.
Nat Commun, 14, 2023
|
|
4D8O
 
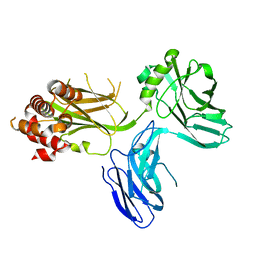 | |
8UQP
 
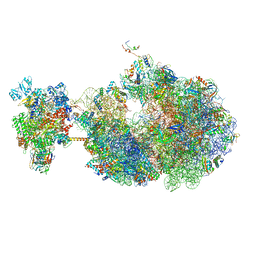 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8URY
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 30 nt long spacer, and fMet-tRNA in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
1PYB
 
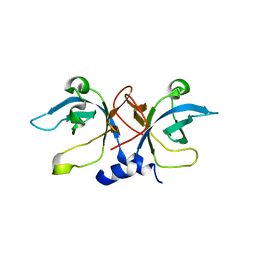 | | Crystal Structure of Aquifex aeolicus Trbp111: a Structure-Specific tRNA Binding Protein | | Descriptor: | tRNA-binding protein Trbp111 | | Authors: | Swairjo, M.A, Morales, A.J, Wang, C.C, Ortiz, A.R, Schimmel, P. | | Deposit date: | 2003-07-08 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of trbp111: a structure-specific tRNA-binding protein.
Embo J., 19, 2000
|
|
6KIN
 
 | | Crystal structure of the tri-functional malyl-CoA lyase from Roseiflexus castenholzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | Authors: | Tang, W.R, Zhang, C.Y, Wang, C, Xu, X.L. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | The C-terminal domain conformational switch revealed by the crystal structure of malyl-CoA lyase from Roseiflexus castenholzii.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
5ZRV
 
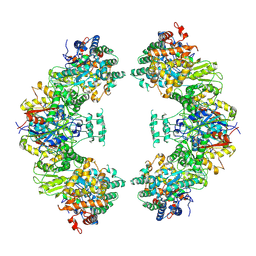 | | Structure of human mitochondrial trifunctional protein, octamer | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Liang, K, Li, N, Dai, J, Wang, X, Liu, P, Chen, X, Wang, C, Gao, N, Xiao, J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM structure of human mitochondrial trifunctional protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZVA
 
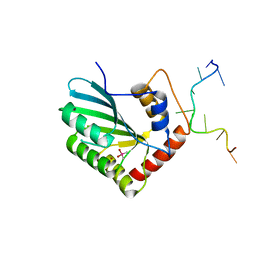 | | APOBEC3F Chimeric Catalytic Domain in Complex with DNA(dC9) | | Descriptor: | APEBEC3F/ssDNA-C9, CACODYLATE ION, DNA (5'-D(*AP*TP*TP*TP*TP*CP*AP*AP*CP*T)-3'), ... | | Authors: | Cheng, C, Zhang, T.L, Wang, C.X, Lan, W.X, Ding, J.P, Cao, C.Y. | | Deposit date: | 2018-05-09 | | Release date: | 2018-11-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Human APOBEC3F Chimeric Catalytic Domain in Complex with DNA
Chin.J.Chem., 36, 2018
|
|
8HSY
 
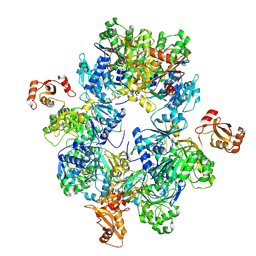 | | Acyl-ACP Synthetase structure | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Acyl-ACP Synthetase structure
To Be Published
|
|
5WZG
 
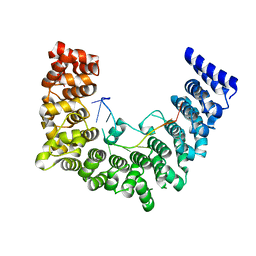 | | Structure of APUM23-GAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
7AKD
 
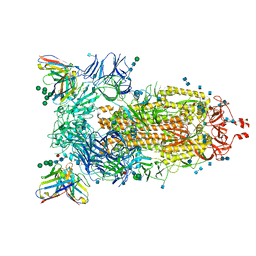 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 47D11 neutralizing antibody heavy chain, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
7AKJ
 
 | | Structure of the SARS-CoV spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-10-01 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
7CWH
 
 | | Structural basis of RACK7 PHD to read a pediatric glioblastoma-associated histone mutation H3.3G34R | | Descriptor: | Peptide from Histone H3.3, Protein kinase C-binding protein 1, ZINC ION | | Authors: | Lan, W.X, Li, Z, Jiao, F.F, Wang, C.X, Guo, R, Cao, C.Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of RACK7 PHD domain to read a pediatric glioblastoma‐associated histone mutation H3.3G34R
Chin.J.Chem., 2021
|
|
5WZJ
 
 | | Structure of APUM23-GGAUUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*UP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5WZH
 
 | | Structure of APUM23-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | Descriptor: | Enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5WZI
 
 | | Structure of APUM23-GGAGUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*GP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
