3BXR
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-N-[(8S,11S)-8-[(1S)-1-methylpropyl]-7,10-dioxo-2-oxa-6,9-diazabicyclo[11.2.2]heptadeca-1(15),13,16-trien-11-yl]-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxamide, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
3BXS
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxylic acid, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
4GN2
 
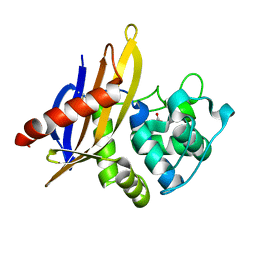 | | Crystal Structure of OXA-45, a Class D beta-lactamase with extended spectrum activity | | Descriptor: | Oxacillinase | | Authors: | Martin, J.D, Xiong, X.L, Catto, L.E, Toleman, M.A, Walsh, T.R, Clarke, A.R, Spencer, J. | | Deposit date: | 2012-08-16 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Kinetic Characterization of OXA-45, a Class D beta-Lactamase with Extended Spectrum activity
To be Published
|
|
2Y8A
 
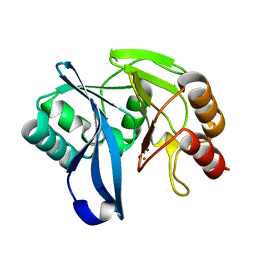 | | VIM-7 with Oxidised. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | MAGNESIUM ION, METALLO-B-LACTAMASE, UNKNOWN ATOM OR ION, ... | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
2Y87
 
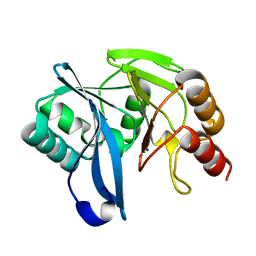 | | Native VIM-7. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | MAGNESIUM ION, METALLO-B-LACTAMASE, UNKNOWN ATOM OR ION, ... | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
5MX9
 
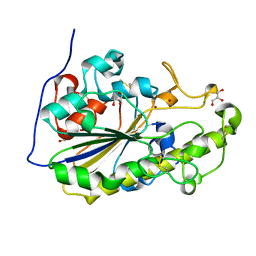 | | High resolution crystal structure of the MCR-2 catalytic domain | | Descriptor: | GLYCEROL, Phosphatidylethanolamine transferase Mcr-2, ZINC ION | | Authors: | Hinchliffe, P, Coates, K, Walsh, T.R, Spencer, J. | | Deposit date: | 2017-01-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 1.12 angstrom resolution crystal structure of the catalytic domain of the plasmid-mediated colistin resistance determinant MCR-2.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2Y8B
 
 | | VIM-7 with Oxidised. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
2FHX
 
 | | Pseudomonas aeruginosa SPM-1 metallo-beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Murphy, T.A, Catto, L.E, Halford, S.E, Hadfield, A.T, Minor, W, Walsh, T.R, Spencer, J. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-17 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pseudomonas aeruginosa SPM-1 Provides Insights into Variable Zinc Affinity of Metallo-beta-lactamases.
J.Mol.Biol., 357, 2006
|
|
4AWY
 
 | | Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, METALLO-BETA-LACTAMASE AIM-1, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
4AX1
 
 | | Q157N mutant. Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | ACETATE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
4AX0
 
 | | Q157A mutant. Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | ACETATE ION, CALCIUM ION, METALLO-BETA-LACTAMASE AIM-1, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157
Antimicrob.Agents Chemother., 56, 2012
|
|
4AWZ
 
 | | Crystal Structure of the Mobile Metallo-beta-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the role of Gln157 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, METALLO-BETA-LACTAMASE AIM-1, ... | | Authors: | Leiros, H.-K.S, Borra, P.S, Brandsdal, B.O, Edvardsen, K.S.W, Spencer, J, Walsh, T.R, Samuelsen, O. | | Deposit date: | 2012-06-06 | | Release date: | 2012-06-20 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Mobile Metallo-Beta-Lactamase Aim-1 from Pseudomonas Aeruginosa: Insights Into Antibiotic Binding and the Role of Gln157.
Antimicrob.Agents Chemother., 56, 2012
|
|
1SQD
 
 | | Structural basis for inhibitor selectivity revealed by crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
1SQI
 
 | | Structural basis for inhibitor selectivity revealed by crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvic acid dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
1SML
 
 | | METALLO BETA LACTAMASE L1 FROM STENOTROPHOMONAS MALTOPHILIA | | Descriptor: | PROTEIN (PENICILLINASE), ZINC ION | | Authors: | Ullah, J.H, Walsh, T.R, Taylor, I.A, Emery, D.C, Verma, C.S, Gamblin, S.J, Spencer, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution.
J.Mol.Biol., 284, 1998
|
|
1TFZ
 
 | | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-05-27 | | Release date: | 2004-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
1TG5
 
 | | Crystal structures of plant 4-hydroxyphenylpyruvate dioxygenases complexed with DAS645 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION, [1-TERT-BUTYL-3-(2,4-DICHLOROPHENYL)-5-HYDROXY-1H-PYRAZOL-4-YL][2-CHLORO-4-(METHYLSULFONYL)PHENYL]METHANONE | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-05-28 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
