9B9M
 
 | | Crystal structure of iron-bound FlcD from Pseudomonas aeruginosa | | Descriptor: | FE (II) ION, Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-04-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
9B9O
 
 | | Crystal structure of FlcD from Pseudomonas aeruginosa bond to iron(II) and substrate | | Descriptor: | (2R)-2-{[(2Z)-2-(hydroxyimino)ethyl]sulfanyl}butanedioic acid, FE (II) ION, Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-04-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
9B9N
 
 | | Crystal structure of FlcD from Pseudomonas aeruginosa bound to iron (II) and substrate | | Descriptor: | (2R)-2-{[(2Z)-2-(hydroxyimino)ethyl]sulfanyl}butanedioic acid, FE (III) ION, Pyrroloquinoline quinone (Coenzyme PQQ) biosynthesis protein C | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-04-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
7KGZ
 
 | |
8ESI
 
 | | Bile Salt Hydrolase from B. longum with covalent inhibitor bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Conjugated bile acid hydrolase | | Authors: | Walker, M.E, Lim, L, Redinbo, M.R. | | Deposit date: | 2022-10-14 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ESG
 
 | | Bile Salt Hydrolase B from Lactobacillus gasseri with covalent inhibitor bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Choloylglycine hydrolase | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-14 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ESL
 
 | | Bile Salt Hydrolase from a Bacteroidales species with covalent inhibitor bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Choloylglycine hydrolase | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-14 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ETK
 
 | | Bile salt hydrolase A from Lactobacillus gasseri bound to covalent probe | | Descriptor: | (5R)-5-[(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-9a,11a-dimethyl-7-(2-{2-[(prop-2-yn-1-yl)oxy]ethoxy}ethoxy)hexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]-1-fluorohexan-2-one (non-preferred name), Conjugated bile salt hydrolase, SODIUM ION | | Authors: | Walker, M.E, Grundy, M.K, Redinbo, M.R. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ETE
 
 | | Bile Salt Hydrolase from B. longum with covalent inhibitor bound | | Descriptor: | (5R)-1-fluoro-5-[(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-7-hydroxy-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]hexan-2-one (non-preferred name), Conjugated bile acid hydrolase | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ETF
 
 | | Bile Salt Hydrolase B from Lactobacillus gasseri with covalent inhibitor bound | | Descriptor: | (5R)-1-fluoro-5-[(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-7-hydroxy-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]hexan-2-one (non-preferred name), Choloylglycine hydrolase, NICKEL (II) ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8EWT
 
 | | Bile salt hydrolase A from Lactobacillus gasseri bound to covalent probe | | Descriptor: | (5R)-5-{(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-9a,11a-dimethyl-7-[(prop-2-yn-1-yl)oxy]hexadecahydro-1H-cyclopenta[a]phenanthren-1-yl}-1-fluorohexan-2-one (non-preferred name), Conjugated bile salt hydrolase, SODIUM ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8W1Q
 
 | | Aerobic crystal structure of iron-bound FlcD from Pseudomonas aeruginosa | | Descriptor: | FE (II) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-02-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
6MXQ
 
 | |
6NOA
 
 | |
7SVH
 
 | | Bile Salt Hydrolase B from Lactobacillus gasseri | | Descriptor: | Choloylglycine hydrolase, MAGNESIUM ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2021-11-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut.
Nat Microbiol, 8, 2023
|
|
7SVG
 
 | |
7SVJ
 
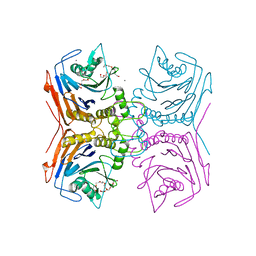 | | Bile Salt Hydrolase from Lactobacillus ingluviei | | Descriptor: | CALCIUM ION, Choloylglycine hydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Walker, M.E, Patel, S, Redinbo, M.R. | | Deposit date: | 2021-11-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut.
Nat Microbiol, 8, 2023
|
|
7SVI
 
 | |
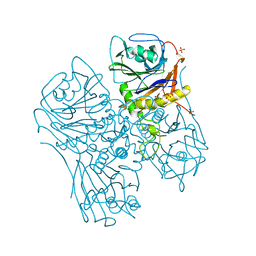
7SVK
 
 | |
7SVE
 
 | |
7SVF
 
 | |
1GPW
 
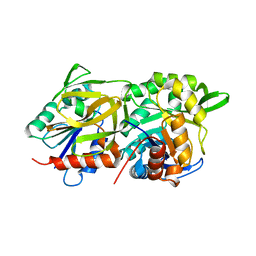 | | Structural evidence for ammonia tunneling across the (beta/alpha)8 barrel of the imidazole glycerol phosphate synthase bienzyme complex. | | Descriptor: | AMIDOTRANSFERASE HISH, HISF PROTEIN, PHOSPHATE ION | | Authors: | Walker, M, Beismann-Driemeyer, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Evidence for Ammonia Tunneling Across the (Beta Alpha)(8) Barrel of the Imidazole Glycerol Phosphate Synthase Bienzyme Complex.
Structure, 10, 2002
|
|
3ZLG
 
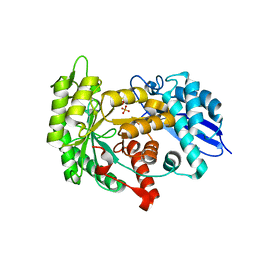 | | Structure of group A Streptococcal enolase K362A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
3ZLH
 
 | | Structure of group A Streptococcal enolase | | Descriptor: | ENOLASE | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
9FJK
 
 | | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6 | | Descriptor: | K501SP6 Fv Heavy Chain, K501SP6 Fv Light Chain, Spike glycoprotein,Fibritin | | Authors: | Bjoernsson, K.H, Walker, M.R, Raghavan, S.S.R, Ward, A.B, Barfod, L.K. | | Deposit date: | 2024-05-31 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6
To Be Published
|
|
