6SK7
 
 | | Cryo-EM structure of rhinovirus-A89 | | Descriptor: | VP1 capsid protein, VP2 capsid protein, VP3 capsid protein, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SK5
 
 | | Cryo-EM structure of rhinovirus-B5 complexed to antiviral OBR-5-340 | | Descriptor: | 6-phenyl-~{N}3-[4-(trifluoromethyl)phenyl]-1~{H}-pyrazolo[3,4-d]pyrimidine-3,4-diamine, Rhinovirus B5 VP1, Rhinovirus B5 VP2, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SK6
 
 | | Cryo-EM structure of rhinovirus-B5 | | Descriptor: | Rhinovirus B5 VP1, Rhinovirus B5 VP2, Rhinovirus B5 VP3, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7PBT
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Wald, J, Fahrenkamp, D, Goessweiner-Mohr, N, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
2WVO
 
 | | Structure of the HET-S N-terminal domain | | Descriptor: | CHLORIDE ION, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Prion Inhibition by Het-S.
Mol.Cell, 38, 2010
|
|
2W75
 
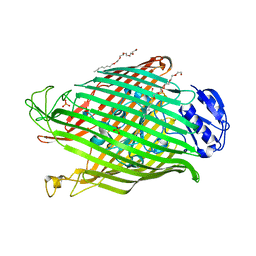 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: Apo-FpvA | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FERRIPYOVERDINE RECEPTOR, PHOSPHATE ION | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W6U
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(G173)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-19 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W6T
 
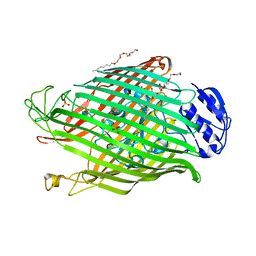 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(DSM50106)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-19 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W77
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(Pfl18.1)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W78
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(ATCC13535)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W76
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(Pa6)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W16
 
 | | Structures of FpvA bound to heterologous pyoverdines | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, DSN-ARG-DSN-FHO-LYS-FHO-THR-THR, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
1S4Y
 
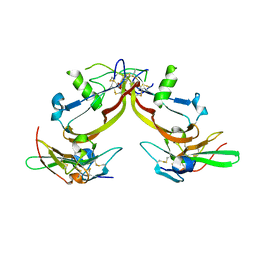 | | Crystal structure of the activin/actrIIb extracellular domain | | Descriptor: | Activin receptor type IIB precursor, Inhibin beta A chain | | Authors: | Greenwald, J, Vega, M.E, Allendorph, G.P, Fischer, W.H, Vale, W, Choe, S, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-01-19 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Flexible Activin Explains the Membrane-Dependent Cooperative Assembly of TGF-beta Family Receptors.
Mol.Cell, 15, 2004
|
|
1LX5
 
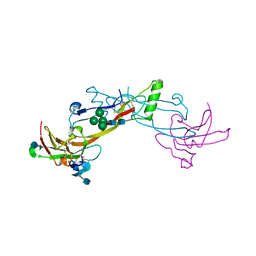 | | Crystal Structure of the BMP7/ActRII Extracellular Domain Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin Type II Receptor, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-04 | | Release date: | 2003-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
1LXI
 
 | | Refinement of BMP7 crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BONE MORPHOGENETIC PROTEIN 7 | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-05 | | Release date: | 2003-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
2WVN
 
 | | Structure of the HET-s N-terminal domain | | Descriptor: | SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The Mechanism of Prion Inhibition by Het-S.
Mol.Cell, 38, 2010
|
|
2WVQ
 
 | | Structure of the HET-s N-terminal domain. Mutant D23A, P33H | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of prion inhibition by HET-S.
Mol. Cell, 38, 2010
|
|
1B9F
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B92
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-19 | | Release date: | 1999-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B9D
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1BTE
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF THE TYPE II ACTIVIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (ACTIVIN RECEPTOR TYPE II) | | Authors: | Greenwald, J, Fischer, W, Vale, W, Choe, S. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase.
Nat.Struct.Biol., 6, 1999
|
|
8C4C
 
 | | F-actin decorated by SipA497-669 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Yuan, B, Wald, J, Marlovits, T.C. | | Deposit date: | 2023-01-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for subversion of host cell actin cytoskeleton during Salmonella infection.
Sci Adv, 9, 2023
|
|
8C4E
 
 | | F-actin decorated by SipA426-685 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Yuan, B, Wald, J, Marlovits, T.C. | | Deposit date: | 2023-01-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for subversion of host cell actin cytoskeleton during Salmonella infection.
Sci Adv, 9, 2023
|
|
6ZNH
 
 | | Structure of the Salmonella PrgI needle filament attached to the basal body | | Descriptor: | PrgI | | Authors: | Lunelli, M, Kotov, V, Wald, J, Kolbe, M, Marlovits, T.C. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical reconstruction of Salmonella and Shigella needle filaments attached to type 3 basal bodies.
Biochem Biophys Rep, 27, 2021
|
|
7AHI
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 2 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
