2IF0
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IEZ
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3VV1
 
 | | Crystal Structure of Caenorhabditis elegans galectin LEC-6 | | Descriptor: | MAGNESIUM ION, Protein LEC-6, beta-D-galactopyranose-(1-4)-alpha-L-fucopyranose | | Authors: | Makyio, H, Takeuchi, T, Tamura, M, Nishiyama, K, Takahashi, H, Natsugari, H, Arata, Y, Kasai, K, Yamada, Y, Wakatsuki, S, Kato, R. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of preferential binding of fucose-containing saccharide by the Caenorhabditis elegans galectin LEC-6
Glycobiology, 23, 2013
|
|
3WAO
 
 | | Crystal structure of Atg13 LIR-fused human LC3B_2-119 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAM
 
 | | Crystal structure of human LC3C_8-125 | | Descriptor: | CITRIC ACID, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAP
 
 | | Crystal structure of Atg13 LIR-fused human LC3C_8-125 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3VHS
 
 | |
3VVH
 
 | |
3VTV
 
 | | Crystal structure of Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3WAL
 
 | | Crystal structure of human LC3A_2-121 | | Descriptor: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3VHT
 
 | | Crystal structure of GFP-Wrnip1 UBZ domain fusion protein in complex with ubiquitin | | Descriptor: | Green fluorescent protein, Green fluorescent protein,ATPase WRNIP1, Ubiquitin, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2011-09-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 2016
|
|
3VTU
 
 | | Crystal structure of human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3VTW
 
 | | Crystal structure of T7-tagged Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3WLU
 
 | | Crystal Structure of human galectin-9 NCRD with Selenolactose | | Descriptor: | 2-(trimethylsilyl)ethyl 4-O-beta-D-galactopyranosyl-6-Se-methyl-6-seleno-beta-D-glucopyranoside, Galectin-9 | | Authors: | Makyio, H, Suzuki, T, Ando, H, Yamada, Y, Ishida, H, Kiso, M, Wakatsuki, S, Kato, R. | | Deposit date: | 2013-11-14 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expanded potential of seleno-carbohydrates as a molecular tool for X-ray structural determination of a carbohydrate-protein complex with single/multi-wavelength anomalous dispersion phasing
Bioorg.Med.Chem., 22, 2014
|
|
3WXA
 
 | | X-ray crystal structural analysis of the complex between ALG-2 and Sec31A peptide | | Descriptor: | Programmed cell death protein 6, Protein transport protein Sec31A, ZINC ION | | Authors: | Takahashi, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2
Int J Mol Sci, 16, 2015
|
|
3WAN
 
 | | Crystal structure of Atg13 LIR-fused human LC3A_2-121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3AXC
 
 | | Crystal structure of linear diubiquitin | | Descriptor: | GLYCEROL, Ubiquitin | | Authors: | Rohaim, A, Kawasaki, M, Kato, R, Dikic, I, Wakatsuki, S. | | Deposit date: | 2011-04-01 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a compact conformation of linear diubiquitin
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2ZAM
 
 | | Crystal structure of mouse SKD1/VPS4B apo-form | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2Z9Z
 
 | | Crystal structure of CERT START domain(N504A mutant), in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZAO
 
 | | Crystal structure of mouse SKD1/VPS4B ADP-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZHM
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 1) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZNE
 
 | | Crystal structure of Zn2+-bound form of des3-23ALG-2 complexed with Alix ABS peptide | | Descriptor: | 16-meric peptide from Programmed cell death 6-interacting protein, Programmed cell death protein 6, SODIUM ION, ... | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZN8
 
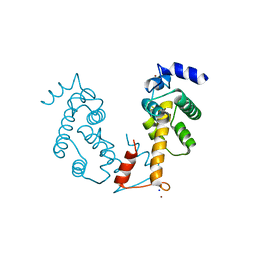 | | Crystal structure of Zn2+-bound form of ALG-2 | | Descriptor: | Programmed cell death protein 6, SODIUM ION, ZINC ION | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZND
 
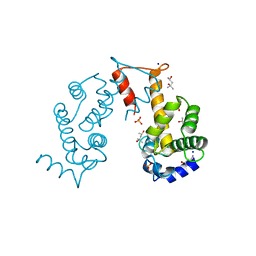 | | Crystal structure of Ca2+-free form of des3-20ALG-2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHOSPHATE ION, Programmed cell death protein 6, ... | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZHN
 
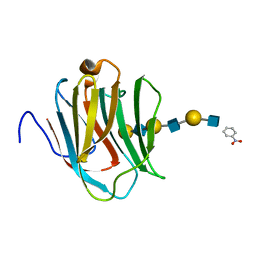 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 2) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, GALECTIN-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
