6TM3
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in a close conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 1,2-ETHANEDIOL, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6YUA
 
 | |
6HR8
 
 | | HMG-CoA reductase from Methanothermococcus thermolithotrophicus in complex with NADPH at 2.9 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, HMG-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wagner, T, Voegeli, B, Erb, T.J, Shima, S. | | Deposit date: | 2018-09-26 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of archaeal HMG-CoA reductase: insights into structural changes of the C-terminal helix of the class-I enzyme.
Febs Lett., 593, 2019
|
|
6HR7
 
 | | HMG-CoA reductase from Methanothermococcus thermolithotrophicus apo form at 2.4 A resolution | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wagner, T, Voegeli, B, Erb, T.J, Shima, S. | | Deposit date: | 2018-09-26 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of archaeal HMG-CoA reductase: insights into structural changes of the C-terminal helix of the class-I enzyme.
Febs Lett., 593, 2019
|
|
5O4J
 
 | | HcgC from Methanococcus maripaludis cocrystallized with SAH and pyridinol | | Descriptor: | (3~{E})-3-[(~{E})-3-oxidanylprop-2-enoyl]iminopropanoic acid, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5L8X
 
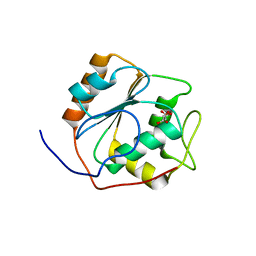 | |
5LAA
 
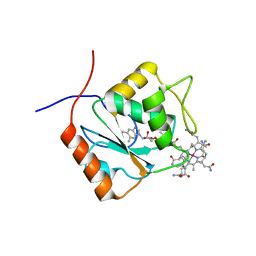 | |
6S6Y
 
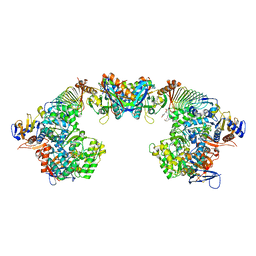 | | X-ray crystal structure of the formyltransferase/hydrolase complex (FhcABCD) from Methylorubrum extorquens in complex with methylofuran | | Descriptor: | (2~{S})-3-[4-[[5-(aminomethyl)furan-3-yl]methoxy]phenyl]-2-(methylamino)propanoic acid, 1,2-ETHANEDIOL, AMINO GROUP, ... | | Authors: | Wagner, T, Hemmann, J.L, Shima, S, Vorholt, J. | | Deposit date: | 2019-07-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5A8K
 
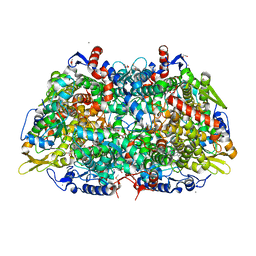 | |
5A8W
 
 | |
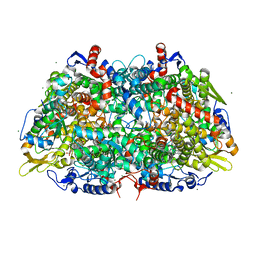
5A8R
 
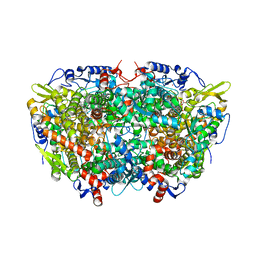 | |
5A0Y
 
 | |
5ODH
 
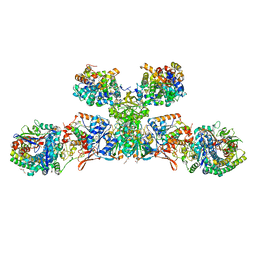 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 3.5 minutes | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
1GG1
 
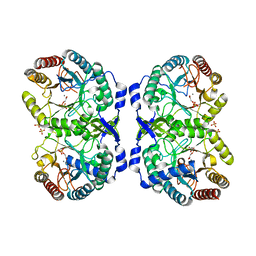 | | CRYSTAL STRUCTURE ANALYSIS OF DAHP SYNTHASE IN COMPLEX WITH MN2+ AND 2-PHOSPHOGLYCOLATE | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE, MANGANESE (II) ION, ... | | Authors: | Wagner, T, Shumilin, I.A, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Escherichia coli: comparison of the Mn(2+)*2-phosphoglycolate and the Pb(2+)*2-phosphoenolpyruvate complexes and implications for catalysis.
J.Mol.Biol., 301, 2000
|
|
5OK4
 
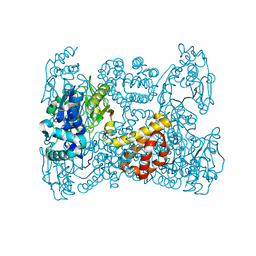 | | Crystal structure of native [Fe]-hydrogenase Hmd from Methanothermobacter marburgensis inactivated by O2. | | Descriptor: | 5'-O-[(S)-{[2-(carboxymethyl)-6-hydroxy-3,5-dimethylpyridin-4-yl]oxy}(hydroxy)phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, FE (III) ION, ... | | Authors: | Wagner, T, Huang, G, Bill, E, Ermler, U, Ataka, K, Shima, S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Dioxygen Sensitivity of [Fe]-Hydrogenase in the Presence of Reducing Substrates.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1GG0
 
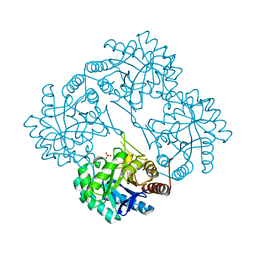 | | CRYSTAL STRUCTURE ANALYSIS OF KDOP SYNTHASE AT 3.0 A | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Wagner, T, Kretsinger, R.H, Bauerle, R, Tolbert, W.D. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3-Deoxy-D-manno-octulosonate-8-phosphate synthase from Escherichia coli. Model of binding of phosphoenolpyruvate and D-arabinose-5-phosphate.
J.Mol.Biol., 301, 2000
|
|
8CNS
 
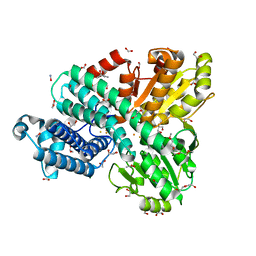 | | The Hybrid Cluster Protein from the thermophilic methanogen Methanothermococcus thermolithotrophicus in a mixed redox state after soaking with hydroxylamine, at 1.36-A resolution. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural and biochemical elucidation of class I hybrid cluster protein natively extracted from a marine methanogenic archaeon.
Front Microbiol, 14, 2023
|
|
8CNR
 
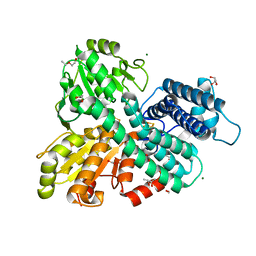 | | Hybrid Cluster Protein from the thermophilic methanogen Methanothermococcus thermolithotrophicus as isolated in a reduced state at 1.45-A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and biochemical elucidation of class I hybrid cluster protein natively extracted from a marine methanogenic archaeon.
Front Microbiol, 14, 2023
|
|
6FRN
 
 | | Structure of F420H2 oxidase (FprA) co-crystallized with 10mM Tb-Xo4 and calcium chloride | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, F420H2 oxidase (FprA), ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Wagner, T, Shima, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2018-02-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
8OOO
 
 | |
6FHL
 
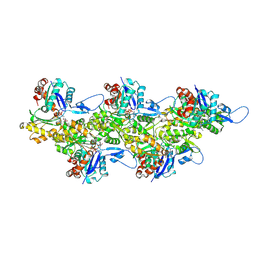 | | Cryo-EM structure of F-actin in complex with ADP-Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Wagner, T, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-06-13 | | Last modified: | 2018-08-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8OOQ
 
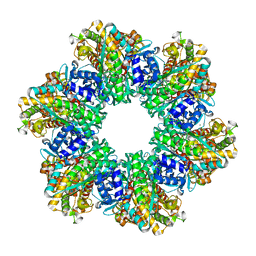 | |
4UUJ
 
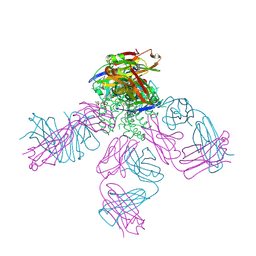 | | POTASSIUM CHANNEL KCSA-FAB WITH TETRAHEXYLAMMONIUM | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Burdette, D, Wagner, T, Focia, P.J, Gross, A. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Kcsa in Complex with Symmetrical Quaternary Ammonium Compounds Reveal a Hydrophobic Binding Site.
Biochemistry, 53, 2014
|
|
8P8G
 
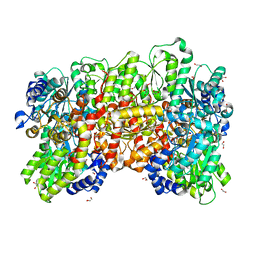 | | Nitrogenase MoFe protein from A. vinelandii beta double mutant D353G/D357G | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Maslac, N, Wagner, T. | | Deposit date: | 2023-06-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Mononuclear Metal-Binding Site of Mo-Nitrogenase Is Not Required for Activity.
Jacs Au, 3, 2023
|
|
8OOL
 
 | |
