9DA5
 
 | | Crystal structure of human DNPH1 bound to inhibitor 2c | | Descriptor: | 5-hydroxymethyl-dUMP N-hydrolase, [(3R,4R)-4-hydroxy-1-{[5-(hydroxymethyl)pyridin-3-yl]methyl}pyrrolidin-3-yl]methyl dihydrogen phosphate | | Authors: | Wagner, A.G, Schramm, V.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Transition State Analogs of Human DNPH1 Reveal Two Electrophile Migration Mechanisms.
J.Med.Chem., 68, 2025
|
|
9DA3
 
 | | Crystal structure of human DNPH1 bound to inhibitor 2a | | Descriptor: | 5-hydroxymethyl-dUMP N-hydrolase, [(3R,4R)-4-hydroxy-1-{[5-(hydroxymethyl)-6-oxo-1,6-dihydropyridin-3-yl]methyl}pyrrolidin-3-yl]methyl dihydrogen phosphate | | Authors: | Wagner, A.G, Schramm, V.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Transition State Analogs of Human DNPH1 Reveal Two Electrophile Migration Mechanisms.
J.Med.Chem., 68, 2025
|
|
9DA1
 
 | | Crystal structure of human DNPH1 bound to inhibitor 1a | | Descriptor: | 5-(hydroxymethyl)uridine 5'-(dihydrogen phosphate), 5-hydroxymethyl-dUMP N-hydrolase | | Authors: | Wagner, A.G, Schramm, V.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Transition State Analogs of Human DNPH1 Reveal Two Electrophile Migration Mechanisms.
J.Med.Chem., 68, 2025
|
|
9DA4
 
 | | Crystal structure of human DNPH1 bound to inhibitor 2b | | Descriptor: | 5-hydroxymethyl-dUMP N-hydrolase, {(3R,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-hydroxypyrrolidin-3-yl}methyl dihydrogen phosphate | | Authors: | Wagner, A.G, Schramm, V.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Transition State Analogs of Human DNPH1 Reveal Two Electrophile Migration Mechanisms.
J.Med.Chem., 68, 2025
|
|
9DA2
 
 | | Crystal structure of human DNPH1 bound to inhibitor 1b | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-deoxy-2-fluoro-5-O-phosphono-beta-D-arabinofuranosyl)-5-(hydroxymethyl)pyrimidine-2,4(1H,3H)-dione, 5-hydroxymethyl-dUMP N-hydrolase, ... | | Authors: | Wagner, A.G, Schramm, V.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Transition State Analogs of Human DNPH1 Reveal Two Electrophile Migration Mechanisms.
J.Med.Chem., 68, 2025
|
|
9DA6
 
 | | Crystal structure of human DNPH1 bound to inhibitor 3a | | Descriptor: | (3R,4R)-3-hydroxy-4-[(phosphonooxy)methyl]pyrrolidinium, 1,2-ETHANEDIOL, 5-hydroxymethyl-dUMP N-hydrolase | | Authors: | Wagner, A.G, Schramm, V.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Transition State Analogs of Human DNPH1 Reveal Two Electrophile Migration Mechanisms.
J.Med.Chem., 68, 2025
|
|
2VJZ
 
 | | Crystal structure form ultalente insulin microcrystals | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Wagner, A, Diez, J, Schulze-Briese, C, Schluckebier, G. | | Deposit date: | 2007-12-14 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Ultralente-A Microcrystalline Insulin Suspension.
Proteins, 74, 2009
|
|
2VK0
 
 | | Crystal structure form ultalente insulin microcrystals | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Wagner, A, Diez, J, Schulze-Briese, C, Schluckebier, G. | | Deposit date: | 2007-12-14 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Ultralente--A Microcrystalline Insulin Suspension.
Proteins, 74, 2009
|
|
4ZG3
 
 | | In-vacuum long-wavelength crystallography | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, SODIUM ION, ... | | Authors: | Wagner, A, Duman, R, Henderson, K, Mykhaylyk, V. | | Deposit date: | 2015-04-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | In-vacuum long-wavelength macromolecular crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6OWY
 
 | | Spy H96L:Im7 K20pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWZ
 
 | | Spy H96L:Im7 L19pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWX
 
 | | Spy H96L:Im7 L18pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8CKM
 
 | | Semaphorin-5A TSR 3-4 domains | | Descriptor: | Semaphorin-5A | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
8CKK
 
 | | Semaphorin-5A TSR 3-4 domains in complex with nitrate | | Descriptor: | NITRATE ION, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
8CKL
 
 | | Semaphorin-5A TSR 3-4 domains in complex with sucrose octasulfate (SOS) | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-2)-1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
9F56
 
 | |
9F5B
 
 | | Identification of zinc ions in LMO4. | | Descriptor: | LIM domain transcription factor LMO4,LIM domain-binding protein 1, ZINC ION | | Authors: | El Omari, K, Forsyth, I, Mancini, E.J, Wagner, A. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Utilizing anomalous signals for element identification in macromolecular crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6FIE
 
 | | Crystallographic structure of calcium loaded Calbindin-D28K. | | Descriptor: | CALCIUM ION, Calbindin, THIOCYANATE ION | | Authors: | Noble, J.W, Almalki, R, Roe, S.M, Wagner, A, Dumanc, R, Atack, J.R. | | Deposit date: | 2018-01-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4Y9O
 
 | | PA3825-EAL Metal-Free-Apo Structure - Manganese Co-crystallisation | | Descriptor: | PA3825-EAL, PHOSPHATE ION | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | MucR and PA3825 EAL-phosphodiesterase domains from Pseudomonas aeruginosa suggest roles for three metals in the active site
To Be Published
|
|
4Y9N
 
 | | PA3825-EAL Metal-Free-Apo Structure - Magnesium Co-crystallisation | | Descriptor: | PA3825-EAL, PHOSPHATE ION | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of PA3825 from P. aeruginosa bound to cyclic di-GMP and pGpG: new insights for a potential three-metal catalytic mechanism of EAL domains
To Be Published
|
|
4Y9M
 
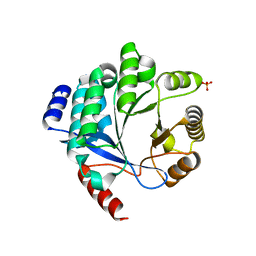 | | PA3825-EAL Metal-Free-Apo Structure | | Descriptor: | PA3825-EAL, PHOSPHATE ION | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
4Y9P
 
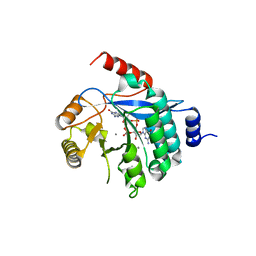 | | PA3825-EAL Ca-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, PA3825-EAL | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | MucR and PA3825 EAL-phosphodiesterase domains from Pseudomonas aeruginosa suggest roles for three metals in the active site
To Be Published
|
|
8PYV
 
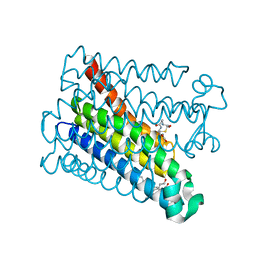 | | Structure of Human PS-1 GSH-analog complex, solved at wavelength 2.755 A | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, Prostaglandin E synthase | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Wagner, A, Vogeley, L, Brown, D.G. | | Deposit date: | 2023-07-26 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX9
 
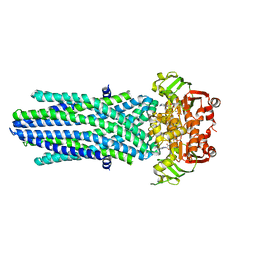 | | Structure of the antibacterial peptide ABC transporter McjD, solved at wavelength 2.75 A | | Descriptor: | MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bountra, K, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PZ4
 
 | | Structure of alginate transporter, AlgE, solved at wavelength 2.755 A | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Wagner, A, Vogeley, L, Brown, D.G. | | Deposit date: | 2023-07-26 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
