8HL3
 
 | |
8HL5
 
 | |
8KAA
 
 | |
8HL4
 
 | |
8HL1
 
 | |
8WQ2
 
 | |
8WQ4
 
 | |
8WKP
 
 | |
5YVE
 
 | |
3DK1
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-0105A | | Descriptor: | (4aR,6r,7aS)-hexahydro-4aH-cyclopenta[b][1,4]dioxin-6-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.F, Weber, I.T. | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Potent HIV-1 protease inhibitors incorporating meso-bicyclic urethanes as P2-ligands: structure-based design, synthesis, biological evaluation and protein-ligand X-ray studies
Org.Biomol.Chem., 6, 2008
|
|
7KQR
 
 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | Authors: | Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
 | |
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
3DJK
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-0255A | | Descriptor: | (5R)-1,3-dioxepan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.F, Weber, I.T. | | Deposit date: | 2008-06-23 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Flexible cyclic ethers/polyethers as novel P2-ligands for HIV-1 protease inhibitors: design, synthesis, biological evaluation, and protein-ligand X-ray studies
J.Med.Chem., 51, 2008
|
|
2F90
 
 | | Crystal structure of bisphosphoglycerate mutase in complex with 3-phosphoglycerate and AlF4- | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, TETRAFLUOROALUMINATE ION | | Authors: | Wang, Y, Liu, L, Wei, Z, Gong, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase
J.Biol.Chem., 281, 2006
|
|
6JPS
 
 | |
8UCR
 
 | |
6IDY
 
 | |
7VGG
 
 | | Cryo-EM structure of Ultraviolet-B activated UVR8 in complex with COP1 | | Descriptor: | E3 ubiquitin-protein ligase COP1, Ultraviolet-B receptor UVR8 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2021-09-16 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into UV-B-activated UVR8 bound to COP1.
Sci Adv, 8, 2022
|
|
2HHJ
 
 | | Human bisphosphoglycerate mutase complexed with 2,3-bisphosphoglycerate (15 days) | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, ... | | Authors: | Wang, Y, Gong, W. | | Deposit date: | 2006-06-28 | | Release date: | 2006-10-24 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase.
J.Biol.Chem., 281, 2006
|
|
3B44
 
 | | Crystal structure of GlpG W136A mutant | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
7E2B
 
 | | GDPD mutant complex from Pyrococcus furiosus DSM 3638 | | Descriptor: | CHOLINE ION, Glycerophosphodiester phosphodiesterase, MAGNESIUM ION, ... | | Authors: | Wang, Y.H, Wang, J, Wang, F.H. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Observation of the GPC production during hydrolysis of LPC by GDPD D60A from Pyrococcus furiosus DSM 3638
To Be Published
|
|
3B45
 
 | | Crystal structure of GlpG at 1.9A resolution | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
7E2A
 
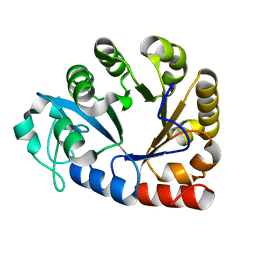 | | GDPD mutant from Pyrococcus furiosus DSM 3638 | | Descriptor: | Glycerophosphodiester phosphodiesterase | | Authors: | Wang, Y.H, Wang, J, Wang, F.H. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of glycerophoshphodiester phospodiesterase D60A mutant from Pyrococcus furiosus DSM
To Be Published
|
|
