7C5L
 
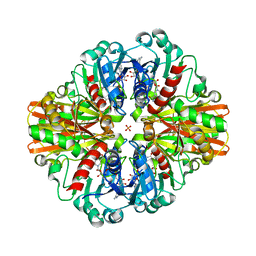 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5P
 
 | | Crystal Structure Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.35 Angstrom resolution. | | Descriptor: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5J
 
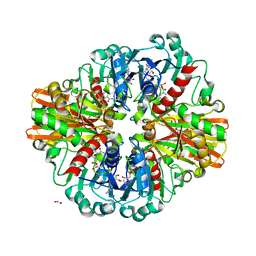 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C7K
 
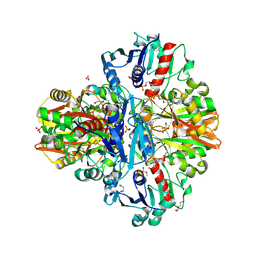 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from Escherichia coli at 1.77 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5O
 
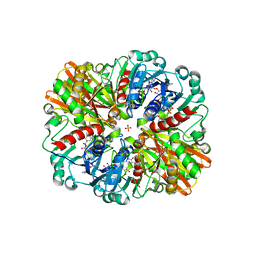 | | Crystal Structure of H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with NAD at 1.98 Angstrom resolution. | | Descriptor: | CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5M
 
 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 1.8 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5G
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7CNO
 
 | | Phomopsin A in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2020-08-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Febs Lett., 595, 2021
|
|
7X4R
 
 | |
6VRG
 
 | | Structure of HIV-1 integrase with native amino-terminal sequence | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Eilers, G, Gupta, K, Allen, A, Zhou, J, Hwang, Y, Cory, M, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2020-02-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influence of the amino-terminal sequence on the structure and function of HIV integrase.
Retrovirology, 17, 2020
|
|
2H7F
 
 | | Structure of variola topoisomerase covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
2H7G
 
 | | Structure of variola topoisomerase non-covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*TP*A)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
1HYV
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH TETRAPHENYL ARSONIUM | | Descriptor: | CHLORIDE ION, INTEGRASE, SULFATE ION, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HYZ
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH A DERIVATIVE OF TETRAPHENYL ARSONIUM. | | Descriptor: | (3,4-DIHYDROXY-PHENYL)-TRIPHENYL-ARSONIUM, CHLORIDE ION, INTEGRASE, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5E6F
 
 | | Canarypox virus resolvase | | Descriptor: | CNPV261 Holliday junction resolvase protein, D(-)-TARTARIC ACID, MAGNESIUM ION | | Authors: | Li, H, Hwang, Y, Perry, K, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Metal Binding Properties of a Poxvirus Resolvase.
J.Biol.Chem., 291, 2016
|
|
1NM3
 
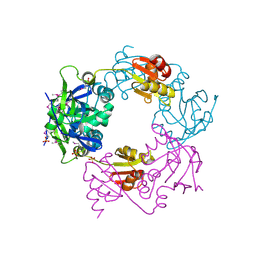 | | Crystal structure of Heamophilus influenza hybrid-Prx5 | | Descriptor: | Protein HI0572, SULFATE ION | | Authors: | Kim, S.J, Woo, J.R, Hwang, Y.S, Jeong, D.G, Shin, D.H, Kim, K.H, Ryu, S.E. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tetrameric Structure of Haemophilus influenza Hybrid Prx5 Reveals Interactions between Electron Donor and Acceptor Proteins.
J.Biol.Chem., 278, 2003
|
|
3IGC
 
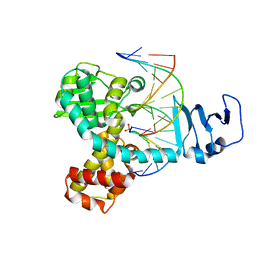 | | Smallpox virus topoisomerase-DNA transition state | | Descriptor: | 5'-D(*AP*TP*TP*CP*C)-3', 5'-D(*CP*GP*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*GP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', ... | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2009-07-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic.
Structure, 18, 2010
|
|
