7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | Descriptor: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CI1
 
 | | Crystal structure of AcrVA2 | | Descriptor: | 1,2-ETHANEDIOL, AcrVA2, SPERMIDINE | | Authors: | Chen, P, Cheng, Z, Wang, Y. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Study on Anti-CRISPR Protein AcrVA2
Prog.Biochem.Biophys., 2021
|
|
4Y0F
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(*GP*TP*TP*GP*AP*GP*CP*GP*TP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Doudeva, L.G, Wang, Y.T, Yuan, H.S. | | Deposit date: | 2015-02-06 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
7CAK
 
 | | SARS-CoV-2 S trimer with three RBD in the open state and complexed with three H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAB
 
 | | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAC
 
 | | SARS-CoV-2 S trimer with one RBD in the open state and complexed with one H014 Fab. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
6J22
 
 | | Crystal structure of Bi-functional enzyme | | Descriptor: | Histidine biosynthesis bifunctional protein HisIE | | Authors: | Zhang, H, Shang, G, Wang, Y. | | Deposit date: | 2018-12-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of Shigella flexneri bi-functional enzyme HisIE in histidine biosynthesis.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6J2L
 
 | | Crystal structure of Bi-functional enzyme | | Descriptor: | Histidine biosynthesis bifunctional protein HisIE, MAGNESIUM ION, ZINC ION | | Authors: | Zhang, H, Shang, G, Wang, Y. | | Deposit date: | 2019-01-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural analysis of Shigella flexneri bi-functional enzyme HisIE in histidine biosynthesis.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
7XAF
 
 | | The crystal structure of TrkA kinase in complex with 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo- tetradecaphan-2-yne-45-carboxamide | | Descriptor: | 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo-tetradecaphan-2-yne-45-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2022-03-17 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001182 Å) | | Cite: | Discovery of the First Highly Selective and Broadly Effective Macrocycle-Based Type II TRK Inhibitors that Overcome Clinically Acquired Resistance.
J.Med.Chem., 65, 2022
|
|
3S54
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug darunavir in space group P21212 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3S53
 
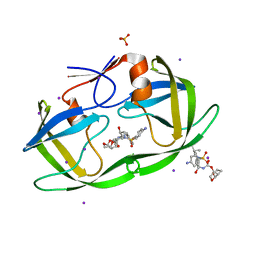 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug darunavir in space group P212121 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, IODIDE ION, PHOSPHATE ION, ... | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
4GKG
 
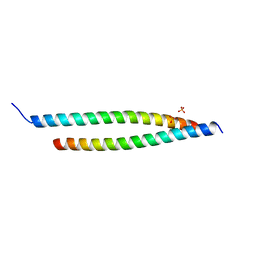 | | Crystal structure of the S-Helix Linker | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, PHOSPHATE ION | | Authors: | Liu, J.W, Lu, D, Sun, Y.J, Wen, J, Yang, Y, Yang, J.G, Wei, X.L, Zhang, X.D, Wang, Y.P. | | Deposit date: | 2012-08-11 | | Release date: | 2013-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of the S-Helix Linker
To be Published
|
|
7XVI
 
 | |
7XVK
 
 | |
7DQ9
 
 | |
8WKC
 
 | | Crystal structure of OgBVMO(Oceanicola granulosus) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Du, Y, Wang, Y.H. | | Deposit date: | 2023-09-27 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of OgBVMO(Oceanicola granulosus)
To be published
|
|
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
7YDX
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound RI-962 | | Descriptor: | 1-methyl-5-[2-(2-methylpropanoylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-phenylethyl]indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Wu, C, Luo, X, Wang, T, Lei, J, Yang, S. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Generative deep learning enables the discovery of a potent and selective RIPK1 inhibitor.
Nat Commun, 13, 2022
|
|
7XZ4
 
 | |
7EBO
 
 | |
6KL5
 
 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
3WHK
 
 | | Crystal structure of PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHL
 
 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
8GUN
 
 | |
8GUO
 
 | |
