8CGF
 
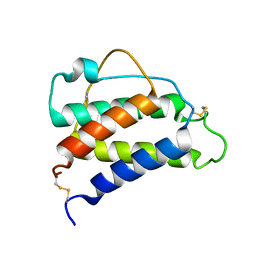 | | Interleukin-4 (wild type) pH 2.4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
7OI3
 
 | | Cryo-EM structure of the Cetacean morbillivirus nucleoprotein-RNA complex | | Descriptor: | Cetacean morbillivirus nucleoprotein, poly-A 6-mer | | Authors: | Zinzula, L, Beck, F, Klumpe, S, Bohn, S, Pfeifer, G, Bollschweiler, D, Nagy, I, Plitzko, J.M, Baumeister, W. | | Deposit date: | 2021-05-11 | | Release date: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the cetacean morbillivirus nucleoprotein-RNA complex.
J.Struct.Biol., 213, 2021
|
|
7O12
 
 | | ABC transporter NosDFY, AMPPNP-bound in GDN | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ABC transporter ATP-binding protein NosF, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O11
 
 | | ABC transporter NosDFY, nucleotide-free in GDN, R-domain 1 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O2U
 
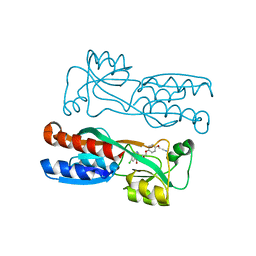 | | PqsR (MvfR) in complex with antagonist 40 | | Descriptor: | 2-[4-[(2S)-3-(6-chloranyl-4-oxidanylidene-quinazolin-3-yl)-2-oxidanyl-propoxy]phenoxy]ethanenitrile, LysR family transcriptional regulator | | Authors: | Emsley, J, Richardson, W. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | PqsR (MvfR) in complex with antagonist 40
To Be Published
|
|
7O10
 
 | | ABC transporter NosDFY, nucleotide-free in GDN, R-domain 2 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O17
 
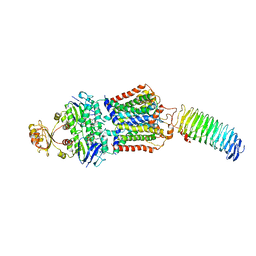 | | ABC transporter NosDFY E154Q, ATP-bound in lipid nanodisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O2T
 
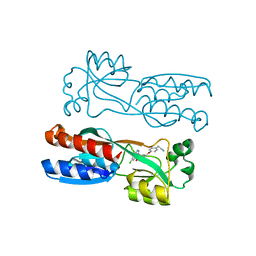 | | PqsR (MvfR) in complex with antagonist 61 | | Descriptor: | 2-[4-[(2S)-3-(6-chloro-4-oxoquinazolin-3-yl)-2-hydroxypropoxy]phenyl]acetonitrile, LysR family transcriptional regulator | | Authors: | Emsley, J, Richardson, W. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | PqsR (MvfR) in complex with antagonist 61
To Be Published
|
|
8CBA
 
 | |
8CCT
 
 | | Crystal structure of the human PXR ligand-binding domain in complex with 2,2'-dichloro bisphenol A | | Descriptor: | 2-chloranyl-4-[2-(3-chloranyl-4-oxidanyl-phenyl)propan-2-yl]phenol, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Derosa, Q, Grimaldi, M, Carivenc, C, Boulahtouf, A, Bourguet, W, Balaguer, P. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the hPXR-LBD in complex with 2,2'-dichloro bisphenol A
To Be Published
|
|
7OC3
 
 | |
1KLK
 
 | | CRYSTAL STRUCTURE OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE TERNARY COMPLEX WITH PT653 AND NADPH | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [N-(2,4-DIAMINOPTERIDIN-6-YL)-METHYL]-DIBENZ[B,F]AZEPINE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Rosowsky, A, Queener, S.F. | | Deposit date: | 2001-12-12 | | Release date: | 2002-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based enzyme inhibitor design: modeling studies and crystal structure analysis of Pneumocystis carinii dihydrofolate reductase ternary complex with PT653 and NADPH.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8CMZ
 
 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CN0
 
 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNA
 
 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
8IDS
 
 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258P in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
8CNC
 
 | | Structure of compound 1 bound KMT9 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein | | Authors: | Sheng, W. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of compound 1 bound KMT9
To Be Published
|
|
8CND
 
 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
8CNB
 
 | | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
5DG4
 
 | | Crystal structure of monomer human cellular retinol binding protein II-Y60L | | Descriptor: | ACETATE ION, Retinol-binding protein 2 | | Authors: | Assar, Z, Nossoni, Z, Wang, W, Gieger, J.H, Borhan, B. | | Deposit date: | 2015-08-27 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
4AIT
 
 | |
2AN7
 
 | | Solution structure of the bacterial antidote ParD | | Descriptor: | Protein parD | | Authors: | Oberer, M, Zangger, K, Gruber, K, Keller, W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ParD, the antidote of the ParDE toxin antitoxin module, provides the structural basis for DNA and toxin binding.
Protein Sci., 16, 2007
|
|
1AQJ
 
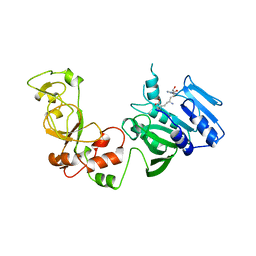 | | STRUCTURE OF ADENINE-N6-DNA-METHYLTRANSFERASE TAQI | | Descriptor: | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI, SINEFUNGIN | | Authors: | Schluckebier, G, Saenger, W. | | Deposit date: | 1996-07-25 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI.
J.Mol.Biol., 265, 1997
|
|
8CNM
 
 | |
8CNO
 
 | |
