1F80
 
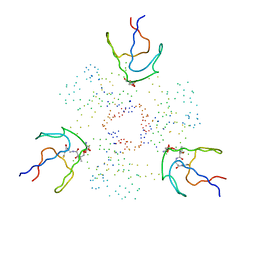 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE IN COMPLEX WITH HOLO-(ACYL CARRIER PROTEIN) | | Descriptor: | ACYL CARRIER PROTEIN, HOLO-(ACYL CARRIER PROTEIN) SYNTHASE, SODIUM ION | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
7XR9
 
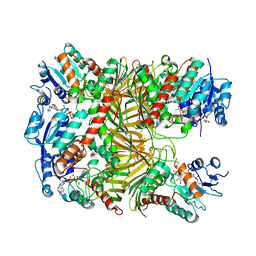 | | Crystal structure of DgpA with glucose | | Descriptor: | DgpA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, beta-D-glucopyranose | | Authors: | Ma, W, He, P. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural mechanism of a dual-functional enzyme DgpA/B/C as both a C -glycoside cleaving enzyme and an O - to C -glycoside isomerase.
Acta Pharm Sin B, 13, 2023
|
|
7YE3
 
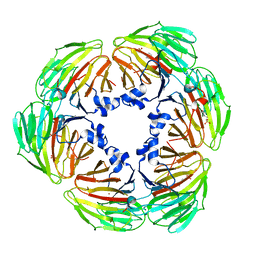 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7YBJ
 
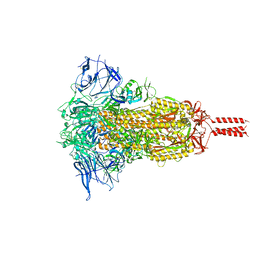 | | SARS-CoV-2 Mu variant spike(close state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBI
 
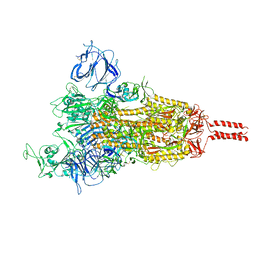 | | SARS-CoV-2 Mu variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YD2
 
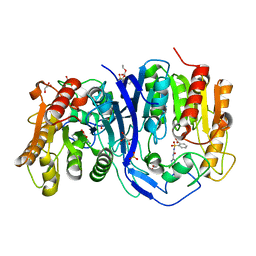 | | SulE_P44R_S209A | | Descriptor: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, ... | | Authors: | Liu, B, Ran, T, Wang, W, He, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7YKP
 
 | | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol | | Descriptor: | GLYCEROL, Triacylglycerol lipase | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase from thermomonospora curvata with glycerol
To Be Published
|
|
7YKO
 
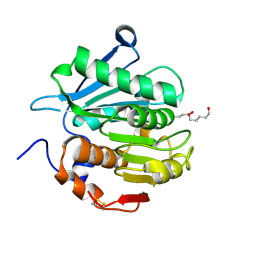 | | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol | | Descriptor: | Triacylglycerol lipase, pentane-1,5-diol | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol
To Be Published
|
|
7YKQ
 
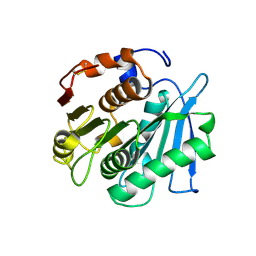 | |
7Y0L
 
 | | SulE-S209A | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | Authors: | Liu, B, Ran, T, He, J, Wang, W. | | Deposit date: | 2022-06-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7YB4
 
 | |
7YBL
 
 | | SARS-CoV-2 B.1.620 variant spike (close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YLQ
 
 | | Crystal structure of Microcystinase C from Sphingomonas sp. ACM-3962 at 2.6 A resolution | | Descriptor: | CALCIUM ION, Microcystinase C, ZINC ION | | Authors: | Guo, X, Li, Z, Ding, W, Yin, P, Feng, L. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Microcystin C from Sphingomonas sp. ACM-3962 at 2.6 A resolution
To Be Published
|
|
7YBH
 
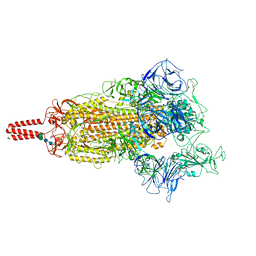 | | SARS-CoV-2 lambda variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBM
 
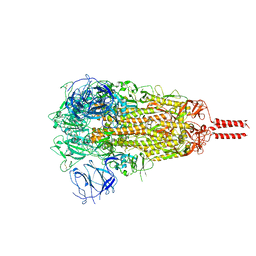 | | SARS-CoV-2 C.1.2 variant spike (Close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBK
 
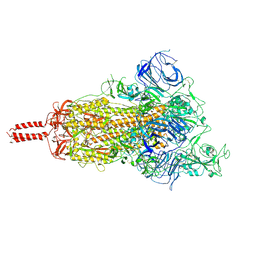 | | SARS-CoV-2 B.1.620 variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7Y53
 
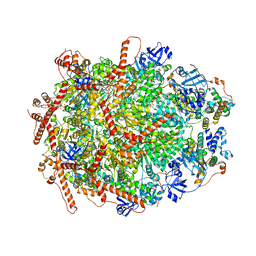 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
1XFR
 
 | |
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1X7B
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH ERB-041 | | Descriptor: | 2-(3-FLUORO-4-HYDROXYPHENYL)-7-VINYL-1,3-BENZOXAZOL-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
