7WJM
 
 | |
7W30
 
 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 1,2-dimethylquinolin-4-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Li, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
7WJO
 
 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
1W9R
 
 | | Solution Structure of Choline Binding Protein A, Domain R2, the Major Adhesin of Streptococcus pneumoniae | | Descriptor: | CHOLINE BINDING PROTEIN A | | Authors: | Luo, R, Mann, B, Lewis, W.S, Rowe, A, Heath, R, Stewart, M.L, Hamburger, A.E, Bjorkman, P.J, Sivakolundu, S, Lacy, E.R, Tuomanen, E, Kriwacki, R.W. | | Deposit date: | 2004-10-15 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Choline Binding Protein A, the Major Adhesin of Streptococcus Pneumoniae
Embo J., 24, 2005
|
|
7WA9
 
 | |
7R35
 
 | | Difference-refined structure of fatty acid photodecarboxylase 300 ns following 400-nm laser irradiation of the dark-state determined by SFX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, W. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WHT
 
 | |
7W5E
 
 | | Oxidase ChaP D49L mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Wang, Y, Zheng, W, Meng, Z, Jin, Y, Zhu, J, Tan, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7RSM
 
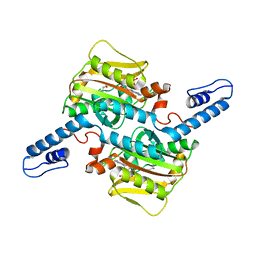 | |
1F46
 
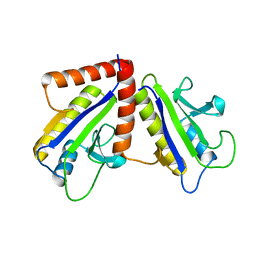 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
1F42
 
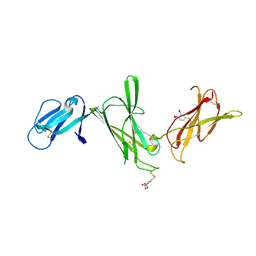 | | THE P40 DOMAIN OF HUMAN INTERLEUKIN-12 | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, INTERLEUKIN-12 BETA CHAIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yoon, C, Johnston, S.C, Tang, J, Tobin, J.F, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12.
EMBO J., 19, 2000
|
|
7XQY
 
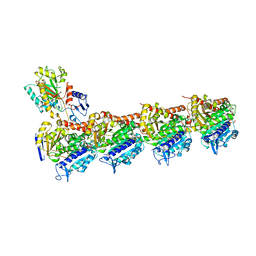 | | Crystal structure of T2R-TTL-15 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-N-(4-methoxyphenyl)-N-methyl-pyrido[3,2-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Lun, T, ChengYong, W. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of T2R-TTL-15 complex
To Be Published
|
|
7XS0
 
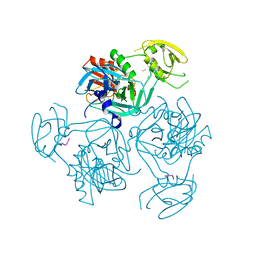 | | Trimer structure of HtrA from Helicobacter pylori bound with a tripeptide | | Descriptor: | Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK | | Authors: | Cui, L, Liu, W. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Int.J.Biol.Macromol., 243, 2023
|
|
6Y6D
 
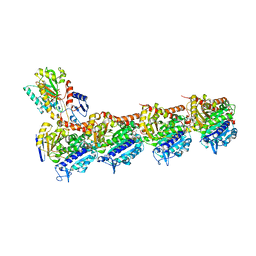 | | Tubulin-7-Aminonoscapine complex | | Descriptor: | (3~{S})-7-azanyl-6-methoxy-3-[(5~{R})-4-methoxy-6-methyl-7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-3~{H}-2-benzofuran-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Oliva, M.A, Prota, A.E, Rodriguez-Salarichs, J, Gu, W, Bennani, Y.L, Jimenez-Barbero, J, Canales, A, Steinmetz, M.O, Diaz, J.F. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Noscapine Activation for Tubulin Binding.
J.Med.Chem., 63, 2020
|
|
7XS2
 
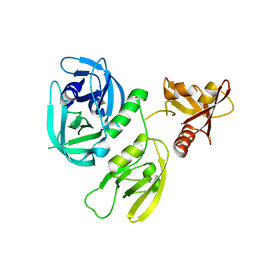 | | Monomer structure of HtrA from Helicobacter pylori | | Descriptor: | Periplasmic serine endoprotease DegP-like | | Authors: | Cui, L, Liu, W. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Int.J.Biol.Macromol., 243, 2023
|
|
7XLC
 
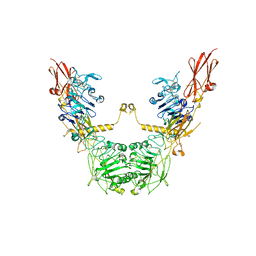 | | conformation II of apo-IGF1R states | | Descriptor: | Insulin-like growth factor 1 receptor | | Authors: | Xi, Z, Cang, W. | | Deposit date: | 2022-04-21 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | conformation II of apo-IGF1R states
To Be Published
|
|
7VMB
 
 | |
7VVN
 
 | | PTH-bound human PTH1R in complex with Gs (class4) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVL
 
 | | PTH-bound human PTH1R in complex with Gs (class2) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VR4
 
 | |
7VR2
 
 | |
7VR0
 
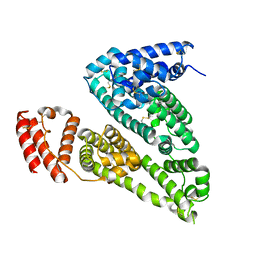 | | Human Serum Albumin | | Descriptor: | Serum albumin | | Authors: | Xiang, W, Yue, Z.L, Su, J.Y. | | Deposit date: | 2021-10-21 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Human Serum Albumin
To Be Published
|
|
7VVK
 
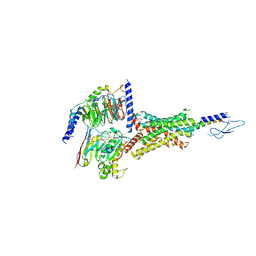 | | PTH-bound human PTH1R in complex with Gs (class1) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVM
 
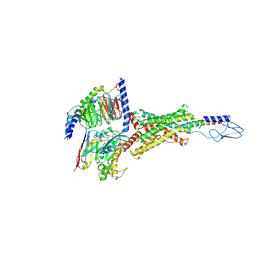 | | PTH-bound human PTH1R in complex with Gs (class3) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VUL
 
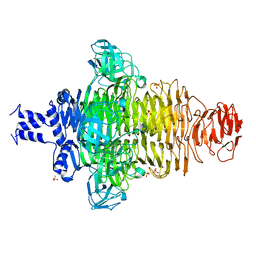 | |
