1LKQ
 
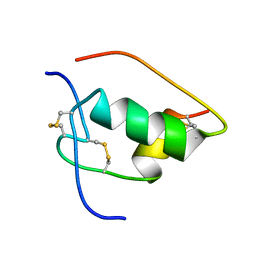 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-GLY, VAL-A3-GLY, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 20 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Weiss, M.A, Hua, Q.X, Chu, Y.C, Jia, W, Philips, N.F, Wang, R.Y, Katsoyannis, P.G. | | Deposit date: | 2002-04-25 | | Release date: | 2002-05-22 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Mechanism of insulin chain combination. Asymmetric roles of A-chain alpha-helices in disulfide pairing
J.Biol.Chem., 277, 2002
|
|
3IZM
 
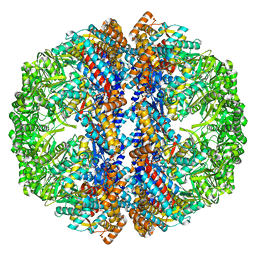 | | Mm-cpn wildtype with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
1UYD
 
 | | Human Hsp90-alpha with 9-Butyl-8-(2-chloro-3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
3IYF
 
 | | Atomic Model of the Lidless Mm-cpn in the Open State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
3J0C
 
 | | Models of E1, E2 and CP of Venezuelan Equine Encephalitis Virus TC-83 strain restrained by a near atomic resolution cryo-EM map | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Zhang, R, Hryc, C.F, Cong, Y, Liu, X, Jakana, J, Gorchakov, R, Baker, M.L, Weaver, S.C, Chiu, W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus.
Embo J., 30, 2011
|
|
3IZN
 
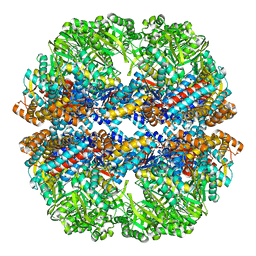 | | Mm-cpn deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3J0F
 
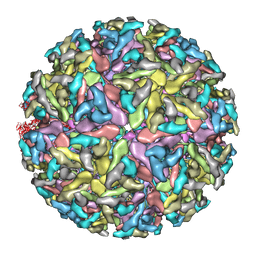 | | Sindbis virion | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Tang, J, Jose, J, Zhang, W, Chipman, P, Kuhn, R.J, Baker, T.S. | | Deposit date: | 2011-07-08 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Molecular Links between the E2 Envelope Glycoprotein and Nucleocapsid Core in Sindbis Virus.
J.Mol.Biol., 414, 2011
|
|
3IZL
 
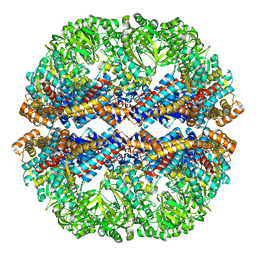 | | Mm-cpn rls deltalid with ATP and AlFx | | Descriptor: | Mm-cpn rls deltalid | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
2FN5
 
 | |
3IZP
 
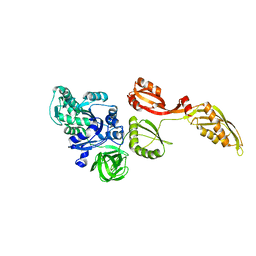 | |
2GM4
 
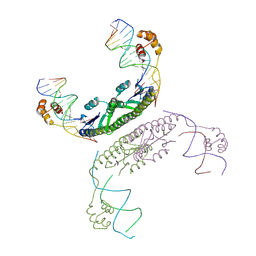 | | An activated, tetrameric gamma-delta resolvase: Hin chimaera bound to cleaved DNA | | Descriptor: | 5'-D(*CP*AP*GP*TP*GP*TP*CP*CP*GP*AP*TP*AP*AP*TP*TP*TP*AP*TP*AP*AP*A)-3', 5'-D(*TP*TP*AP*TP*CP*GP*GP*AP*CP*AP*CP*TP*G)-3', Transposon gamma-delta resolvase | | Authors: | Kamtekar, S, Ho, R.S, Li, W, Steitz, T.A. | | Deposit date: | 2006-04-05 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Implications of structures of synaptic tetramers of gamma delta resolvase for the mechanism of recombination.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3O40
 
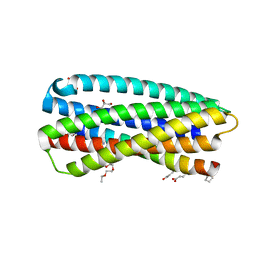 | | Complex of a chimeric alpha/beta-peptide based on the gp41 CHR domain bound to gp41-5 | | Descriptor: | CHLORIDE ION, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2010-07-26 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad Distribution of Energetically Important Contacts across an Extended Protein Interface.
J.Am.Chem.Soc., 133, 2011
|
|
3IZK
 
 | | Mm-cpn rls deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
1UY8
 
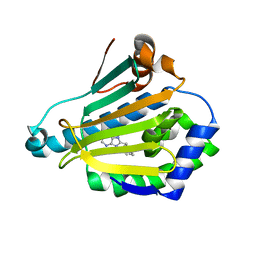 | | Human Hsp90-alpha with 9-Butyl-8-(3-trimethoxy-benzyl)-9H-purin-6ylamine | | Descriptor: | 9-BUTYL-8-(3-METHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
4MB4
 
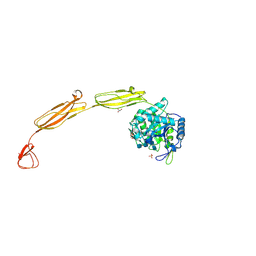 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3O42
 
 | |
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
1UMS
 
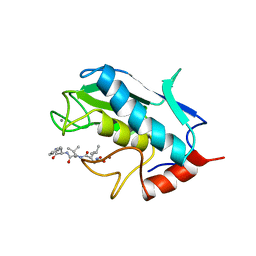 | | STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
3IGO
 
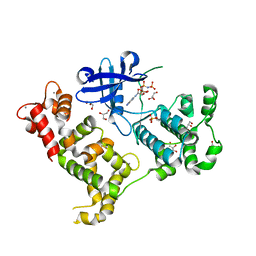 | | Crystal structure of Cryptosporidium parvum CDPK1, cgd3_920 | | Descriptor: | CALCIUM ION, Calmodulin-domain protein kinase 1, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Amani, M, Allali-Hassanali, A, Vedadi, M, Tempel, W, MacKenzie, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3III
 
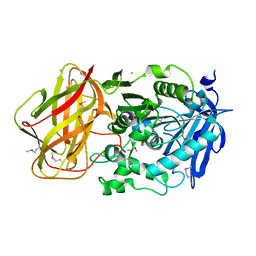 | | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Osinski, T, Chruszcz, M, Domagalski, M.J, Cymborowski, M, Shumilin, I.A, Skarina, T, Onopriyenko, O, Zimmerman, M.D, Savchenko, A, Edwards, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-01 | | Release date: | 2009-08-18 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus
To be Published
|
|
3IVZ
 
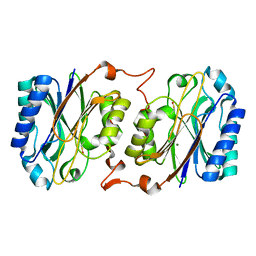 | | Crystal structure of hyperthermophilic nitrilase | | Descriptor: | MAGNESIUM ION, Nitrilase | | Authors: | Raczynska, J, Vorgias, C, Antranikian, G, Rypniewski, W. | | Deposit date: | 2009-09-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic analysis of a thermoactive nitrilase.
J.Struct.Biol., 173, 2010
|
|
2PM1
 
 | |
2PMB
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Zhan, C, Shi, W, Toro, R, Sauder, J.M, Gilmore, J, Iizuka, M, Maletic, M, Gheyi, T, Wasserman, S.R, Smith, D, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Vibrio Cholerae.
To be Published
|
|
3IWH
 
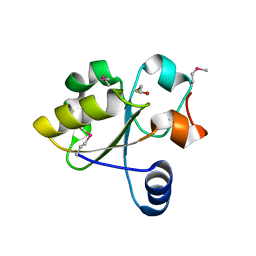 | | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus | | Descriptor: | BETA-MERCAPTOETHANOL, Rhodanese-like domain protein | | Authors: | Kim, Y, Chruszcz, M, Minor, W, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus
To be Published
|
|
2GLP
 
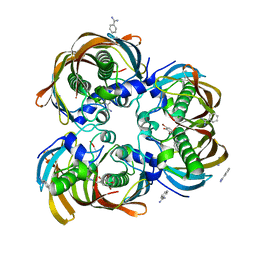 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with compound 1 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
