3IZJ
 
 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3J0G
 
 | |
1CVI
 
 | | CRYSTAL STRUCTURE OF HUMAN PROSTATIC ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Jakob, C.G, Lewinski, K, Kuciel, R, Ostrowski, W, Lebioda, L. | | Deposit date: | 1999-08-23 | | Release date: | 1999-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human prostatic acid phosphatase .
Prostate, 42, 2000
|
|
4YVD
 
 | | Crytsal structure of human Pleiotropic Regulator 1 (PRL1) | | Descriptor: | CHLORIDE ION, Pleiotropic regulator 1, SODIUM ION, ... | | Authors: | Dong, A, Zeng, H, Xu, C, Tempel, W, Li, Y, He, H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crytsal structure of human Pleiotropic Regulator 1 (PRL1).
to be published
|
|
2VSI
 
 | | Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, 4-AMINO-1-{5-O-[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]-ALPHA-D-ARABINOFURANOSYL}PYRIMIDIN-2(1H)-ONE, CALCIUM ION | | Authors: | Baur, S, Marles-Wright, J, Buckenmaier, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthesis of Cdp-Activated Ribitol for Teichoic Acid Precursors in Streptococcus Pneumoniae.
J.Bacteriol., 191, 2009
|
|
3G70
 
 | | Design and Preparation of Potent, Non-Peptidic, Bioavailable Renin Inhibitors | | Descriptor: | (1R,5S)-7-{4-[3-(2-chloro-3,6-difluorophenoxy)propyl]phenyl}-N-cyclopropyl-N-(2,3-dichlorobenzyl)-3,9-diazabicyclo[3.3.1]non-6-ene-6-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Bezencon, O, Bur, D, Prade, L, Weller, T, Boss, C, Fischli, W. | | Deposit date: | 2009-02-09 | | Release date: | 2009-06-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Preparation of Potent, Nonpeptidic, Bioavailable Renin Inhibitors
J.Med.Chem., 52, 2009
|
|
1DMK
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 4-AMINO-6-PHENYL-TETRAHYDROPTERIDINE | | Descriptor: | 2,4-DIAMINO-6-PHENYL-5,6,7,8,-TETRAHYDROPTERIDINE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
6IO9
 
 | | The structure of apo-UdgX | | Descriptor: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, SULFATE ION | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
6IOB
 
 | | The structure of the H109A mutant of UdgX in complex with uracil | | Descriptor: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, URACIL | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
1YG2
 
 | | Structure of the Vibrio cholerae virulence activator AphA | | Descriptor: | gene activator AphA | | Authors: | De Silva, R.S, Kovacikova, G, Lin, W, Taylor, R.T, Skorupski, K, Kull, F.J. | | Deposit date: | 2005-01-04 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily
J.Biol.Chem., 280, 2005
|
|
4WW9
 
 | |
3GBJ
 
 | | Crystal structure of the motor domain of kinesin KIF13B bound with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF13B protein, MAGNESIUM ION, ... | | Authors: | Tong, Y, Shen, L, Shen, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of the motor domain of kinesin KIF13B bound with ADP
To be Published
|
|
4WYU
 
 | |
7S9E
 
 | | Cryo-EM Structure of dolphin Prestin: Inhibited II (Sulfate +Salicylate) state | | Descriptor: | 2-HYDROXYBENZOIC ACID, Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S8X
 
 | | Cryo-EM Structure of dolphin Prestin: Sensor Up (compact) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9C
 
 | | Cryo-EM Structure of dolphin Prestin: Sensor Down II (Expanded II) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9D
 
 | | Cryo-EM Structure of dolphin Prestin: Intermediate state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
4GGC
 
 | |
1DP9
 
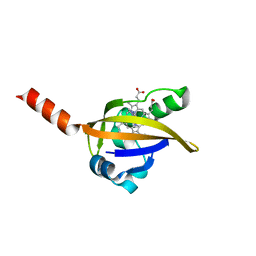 | | CRYSTAL STRUCTURE OF IMIDAZOLE-BOUND FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-24 | | Release date: | 2000-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
2RRT
 
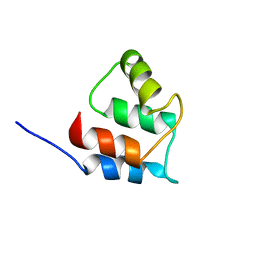 | |
4X3X
 
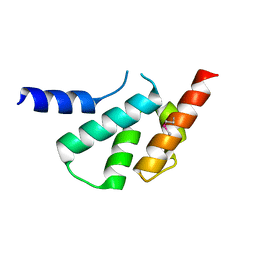 | | The crystal structure of Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
7S9A
 
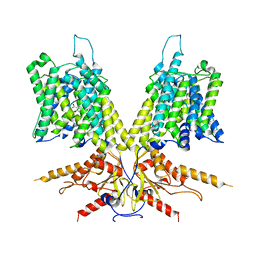 | | Cryo-EM Structure of dolphin Prestin: Inhibited I (Chloride + Salicylate) | | Descriptor: | 2-HYDROXYBENZOIC ACID, Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9B
 
 | | Cryo-EM Structure of dolphin Prestin: Sensor Down I (Expanded) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
1Y4K
 
 | | Lipoxygenase-1 (Soybean) at 100K, N694G Mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (II) ION, ... | | Authors: | Chruszcz, M, Segraves, E, Holman, T.R, Minor, W. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic, spectroscopic, and structural investigations of the soybean lipoxygenase-1 first-coordination sphere mutant, Asn694Gly.
Biochemistry, 45, 2006
|
|
1YFD
 
 | | Crystal structure of the Y122H mutant of ribonucleotide reductase R2 protein from E. coli | | Descriptor: | MERCURY (II) ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Kolberg, M, Logan, D.T, Bleifuss, G, Poetsch, S, Sjoeberg, B.M, Graeslund, A, Lubitz, W, Lassmann, G, Lendzian, F. | | Deposit date: | 2004-12-31 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new tyrosyl radical on Phe208 as ligand to the diiron center in Escherichia coli ribonucleotide reductase, mutant R2-Y122H. Combined x-ray diffraction and EPR/ENDOR studies
J.Biol.Chem., 280, 2005
|
|
