7GP1
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z401437758 | | Descriptor: | 1-(2-methylphenyl)-1,2,3-triazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GOA
 
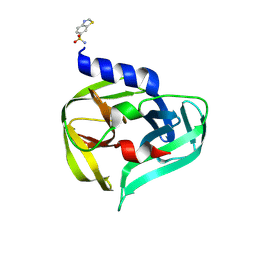 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1269184613 | | Descriptor: | 1,3-benzothiazole-6-sulfonamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GP3
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z437516460 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(6-methylpyridin-3-yl)methyl]cyclobutanecarboxamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GOE
 
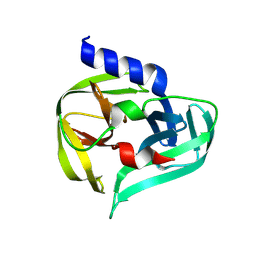 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1342868616 | | Descriptor: | 1-(diphenylmethyl)azetidin-3-ol, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GP4
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z44585777 | | Descriptor: | DIMETHYL SULFOXIDE, N-phenylpyrrolidine-1-carboxamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GOU
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z285782452 | | Descriptor: | N-methyl-2-(methylsulfonyl)aniline, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GP7
 
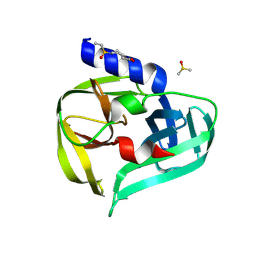 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z53825020 | | Descriptor: | 1-(ethanesulfonyl)piperidine-4-carboxylic acid, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GOW
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z31432964 | | Descriptor: | (4-methylpiperazin-1-yl)[(2R)-oxolan-2-yl]methanone, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GPA
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z56978034 | | Descriptor: | (4-fluorophenoxy)acetic acid, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7KRS
 
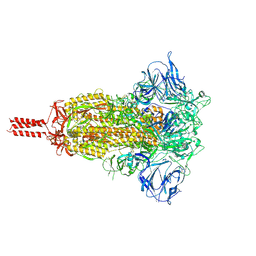 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7KRQ
 
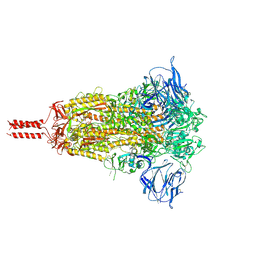 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
1ALK
 
 | |
7KZI
 
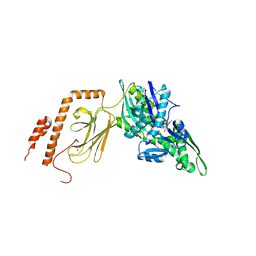 | | Intermediate state (QQQ) of near full-length DnaK alternatively fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7KZU
 
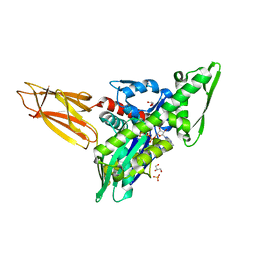 | | Quasi-intermediate state (Q) of a truncated Hsp70 DnaK fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, GLYCEROL, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7RND
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VDPVD-CHO | | Descriptor: | Ac-VDPVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7RN8
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VD(Orn)VD-CHO | | Descriptor: | Ac-VD(Orn)VD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7KW4
 
 | | L-DNA containing 2'-fluoro modification | | Descriptor: | DNA (5'-D(*(0DG)P*(0FU)P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DA)P*(0DC))-3'), MAGNESIUM ION | | Authors: | Zhang, W, Zhang, Y. | | Deposit date: | 2020-11-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Synthesis and Structural Characterization of 2'-Deoxy-2'-fluoro-l-uridine Nucleic Acids.
Org.Lett., 23, 2021
|
|
7RNE
 
 | | Crystal structure of caspase-3 with inhibitor Ac-YKPVD-CHO | | Descriptor: | Ac-YKPVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7RM9
 
 | | Human Malate Dehydrogenase I (MDHI) | | Descriptor: | MALONATE ION, Malate dehydrogenase, cytoplasmic | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of the Human Cytosolic Malate Dehydrogenase I.
Acs Omega, 7, 2022
|
|
7RN7
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VD(Aly)VD-CHO | | Descriptor: | Ac-VD(Aly)VD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7KRW
 
 | |
7RNB
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VDRVD-CHO | | Descriptor: | Ac-VDRVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7KX0
 
 | | Crystal structure of the CD27:CD70 co-stimulatory complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD27 antigen, ... | | Authors: | Maben, Z, Liu, W, Mosyak, L, Chaparro-Riggers, J. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural delineation and phase-dependent activation of the costimulatory CD27:CD70 complex.
J.Biol.Chem., 297, 2021
|
|
7KO2
 
 | |
7RN9
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VDFVD-CHO | | Descriptor: | Ac-VDFVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
