1UNC
 
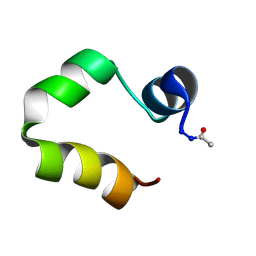 | | Solution structure of the human villin C-terminal headpiece subdomain | | Descriptor: | VILLIN 1 | | Authors: | Vermeulen, W, Van Troys, M, Vanhaesebrouck, P, Verschueren, M, Fant, F, Ampe, C, Martins, J, Borremans, F. | | Deposit date: | 2003-09-09 | | Release date: | 2004-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the C-Terminal Headpiece Subdomains of Human Villin and Advillin, Evaluation of Headpiece F-Actin-Binding Requirements
Protein Sci., 13, 2004
|
|
2O1O
 
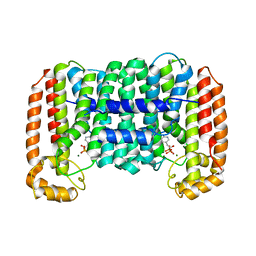 | | Cryptosporidium parvum putative polyprenyl pyrophosphate synthase (cgd4_2550) in complex with risedronate. | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, MAGNESIUM ION, Putative farnesyl pyrophosphate synthase | | Authors: | Chruszcz, M, Artz, J.D, Dong, A, Dunford, J, Lew, J, Zhao, Y, Kozieradski, I, Kavanaugh, K.L, Oppermann, U, Sundstrom, M, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Minor, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Targeting a uniquely nonspecific prenyl synthase with bisphosphonates to combat cryptosporidiosis
Chem.Biol., 15, 2008
|
|
5TO2
 
 | |
4BWJ
 
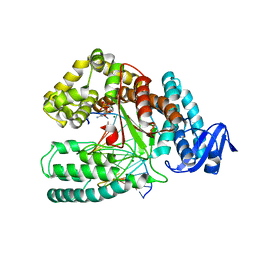 | | KlenTaq mutant in complex with DNA and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOCP)-3', ... | | Authors: | Blatter, N, Bergen, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-07-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and Function of an RNA-Reading Thermostable DNA Polymerase.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1XM1
 
 | | Nonbasic Thrombin Inhibitor Complex | | Descriptor: | Hirudin, N-{[(2S)-1-(N-{[4-({[AMINO(IMINO)METHYL]AMINO}METHYL)CYCLOHEXYL]CARBONYL}-3-CYCLOHEXYL-L-ALANYL)AZETIDIN-2-YL]CARBONYL}-L-TYROSYL-N~6~-[AMINO(IMINO)METHYL]-L-LYSINAMIDE, thrombin | | Authors: | Friedrich, R, Bode, W, Schwienhorst, A. | | Deposit date: | 2004-10-01 | | Release date: | 2005-05-10 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nonbasic Thrombin Inhibitor Complex
To be Published
|
|
4QGL
 
 | | Acireductone dioxygenase from Bacillus anthracis with three cadmium ions | | Descriptor: | Acireductone dioxygenase, CADMIUM ION | | Authors: | Milaczewska, A.M, Chruszcz, M, Majorek, K.A, Porebski, P.J, Borowski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Acireductone dioxygenase from Bacillus anthracis with three cadmium ions
To be Published
|
|
4KH1
 
 | | The R state structure of E. coli ATCase with CTP,UTP, and Magnesium bound | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
3NEY
 
 | | Crystal structure of the kinase domain of MPP1/p55 | | Descriptor: | 55 kDa erythrocyte membrane protein, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tong, Y, Zhong, N, Guan, X, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the kinase domain of MPP1/p55
To be Published
|
|
2OBM
 
 | | Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, EscN | | Authors: | Zarivach, R, Vuckovic, M, Deng, W, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of a prototypical ATPase from the type III secretion system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2OCB
 
 | | Crystal structure of human RAB9B in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-9B, ... | | Authors: | Hong, B, Shen, L, Walker, J.R, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human RAB9B in complex with a GTP analogue
To be Published
|
|
1ZOB
 
 | |
4QP4
 
 | |
1UJD
 
 | | Solution Structure of RSGI RUH-003, a PDZ domain of hypothetical KIAA0559 protein from human cDNA | | Descriptor: | KIAA0559 protein | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-003, a PDZ domain of hypothetical KIAA0559 protein from human cDNA
To be published
|
|
4QPA
 
 | |
1RQL
 
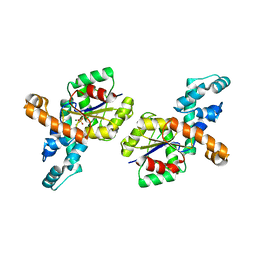 | | Crystal Structure of Phosponoacetaldehyde Hydrolase Complexed with Magnesium and the Inhibitor Vinyl Sulfonate | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase, VINYLSULPHONIC ACID | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
2OEN
 
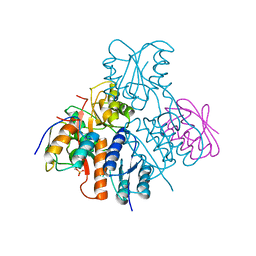 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose-6-phosphate and fructose-1,6-bisphosphate | | Descriptor: | Catabolite control protein, Phosphocarrier protein HPr | | Authors: | Schumacher, M.A, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-12-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
3NA8
 
 | | Crystal Structure of a putative dihydrodipicolinate synthetase from Pseudomonas aeruginosa | | Descriptor: | D-MALATE, MAGNESIUM ION, putative dihydrodipicolinate synthetase | | Authors: | Qiu, W, Lam, R, Romanov, V, Jones, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-06-01 | | Release date: | 2011-06-01 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a putative dihydrodipicolinate synthetase from Pseudomonas aeruginosa
To be Published
|
|
1RGL
 
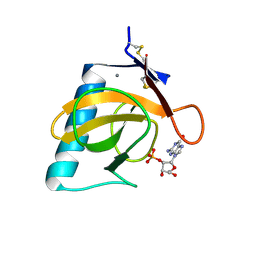 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
2OCP
 
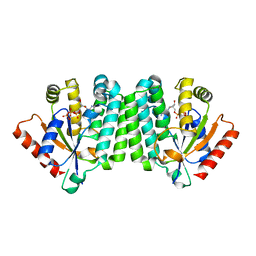 | | Crystal Structure of Human Deoxyguanosine Kinase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxyguanosine kinase | | Authors: | Johansson, K, Ramaswamy, S, Ljungkrantz, C, Knecht, W, Piskur, J, Munch-Petersen, B, Eriksson, S, Eklund, H. | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Substrate Specificities of Cellular Deoxyribonucleoside Kinases.
Nat.Struct.Biol., 8, 2001
|
|
1Z09
 
 | | Solution structure of km23 | | Descriptor: | Dynein light chain 2A, cytoplasmic | | Authors: | Ilangovan, U, Ding, W, Mulder, K, Hinck, A.P, Zuniga, J, Trbovich, J.A, Demeler, B, Groppe, J.C. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Homodimeric Dynein Light Chain km23.
J.Mol.Biol., 352 (2), 2005
|
|
2OK5
 
 | | Human Complement factor B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement factor B, ... | | Authors: | Milder, F.J, Gomes, L, Schouten, A, Janssen, B.J.C, Huizinga, E.G, Romijn, R.A, Hemrika, W, Roos, A, Daha, M.R, Gros, P. | | Deposit date: | 2007-01-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Factor B structure provides insights into activation of the central protease of the complement system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4QC2
 
 | | Crystal structure of lipopolysaccharide transport protein LptB in complex with ATP and Magnesium ions | | Descriptor: | ABC transporter related protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, Z, Xiang, Q, Zhu, X, Dong, H, He, C, Wang, H, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
2OGP
 
 | | Solution structure of the second PDZ domain of Par-3 | | Descriptor: | Partitioning-defective 3 homolog | | Authors: | Feng, W, Wu, H, Chen, J, Chan, L.-N, Zhang, M. | | Deposit date: | 2007-01-07 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | PDZ domains of par-3 as potential phosphoinositide signaling integrators
Mol.Cell, 28, 2007
|
|
3NOJ
 
 | | The structure of HMG/CHA aldolase from the protocatechuate degradation pathway of Pseudomonas putida | | Descriptor: | 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Kimber, M.S, Wang, W, Mazurkewich, S, Seah, S.Y.K. | | Deposit date: | 2010-06-25 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Kinetic Characterization of 4-Hydroxy-4-methyl-2-oxoglutarate/4-Carboxy-4-hydroxy-2-oxoadipate Aldolase, a Protocatechuate Degradation Enzyme Evolutionarily Convergent with the HpaI and DmpG Pyruvate Aldolases.
J.Biol.Chem., 285, 2010
|
|
4QEI
 
 | | Two distinct conformational states of GlyRS captured in crystal lattice | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine--tRNA ligase, tRNA-Gly-CCC-2-2 | | Authors: | Xie, W, Qin, X, Deng, X, Zhang, Q, Li, Q. | | Deposit date: | 2014-05-16 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
