3GN5
 
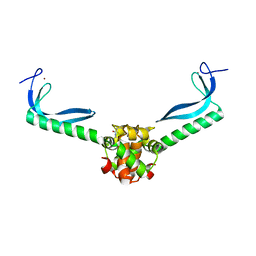 | | Structure of the E. coli protein MqsA (YgiT/b3021) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator MQSA (YGIT/b3021), ZINC ION | | Authors: | Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | Deposit date: | 2009-03-16 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
1KM1
 
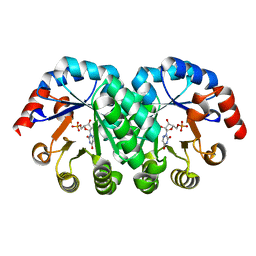 | |
2OLK
 
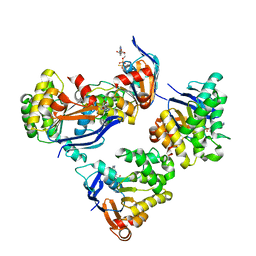 | | ABC Protein ArtP in complex with ADP-beta-S | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, Amino acid ABC transporter | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the ATP-binding cassette (ABC) protein ArtP from Geobacillus stearothermophilus reveal a stable dimer in the post hydrolysis state and an asymmetry in the dimerization region
To be Published
|
|
3M7I
 
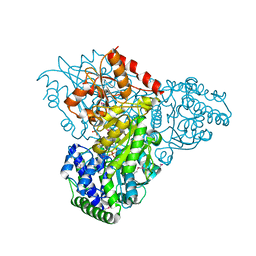 | | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate(pyranose form) and magnesium ion | | Descriptor: | 1,2-ETHANEDIOL, 5-O-phosphono-beta-D-ribofuranose, MAGNESIUM ION, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of transketolase in complex with thiamine diphosphate, ribose-5-phosphate(pyranose form) and magnesium ion
TO BE PUBLISHED
|
|
7BV8
 
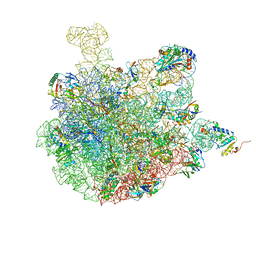 | | Mature 50S ribosomal subunit from RrmJ knock out E.coli strain | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Wang, W, Li, W.Q, Ge, X.L, Yan, K.G, Mandava, C.S, Sanyal, S, Gao, N. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Loss of a single methylation in 23S rRNA delays 50S assembly at multiple late stages and impairs translation initiation and elongation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2OM5
 
 | |
6D0S
 
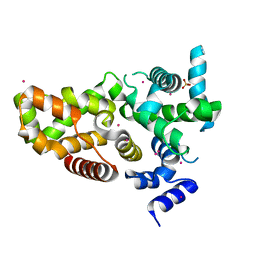 | | RabGAP domain of human TBC1D22B | | Descriptor: | SULFATE ION, TBC1 domain family member 22B, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RabGAP domain of human TBC1D22B
To be Published
|
|
2H2S
 
 | |
5IIU
 
 | | Crystal structure of Equine Serum Albumin in the presence of 10 mM zinc at pH 6.9 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Cymborowski, M.T, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
2OV5
 
 | | Crystal structure of the KPC-2 carbapenemase | | Descriptor: | BICINE, Carbapenemase | | Authors: | Ke, W, Bethel, C.R, Thomson, J.M, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of KPC-2: Insights into Carbapenemase Activity in Class A beta-Lactamases.
Biochemistry, 46, 2007
|
|
1U1E
 
 | | Structure of e. coli uridine phosphorylase complexed to 5(phenylseleno)acyclouridine (PSAU) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(PHENYLSELANYL)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2OWA
 
 | | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040) | | Descriptor: | Arfgap-like finger domain containing protein, ZINC ION | | Authors: | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040)
To be Published
|
|
8PFC
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2H67
 
 | | NMR structure of human insulin mutant HIS-B5-ALA, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hua, Q.X, Liu, M, Hu, S.Q, Jia, W, Arvan, P, Weiss, M.A. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | A Conserved Histidine in Insulin Is Required for the Foldability of Human Proinsulin: Structure and function of an Alab5 analog.
J.Biol.Chem., 281, 2006
|
|
3ET3
 
 | |
1TW0
 
 | |
1U3P
 
 | | IspF native | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1TX7
 
 | | Bovine Trypsin complexed with p-amidinophenylmethylphosphinic acid (AMPA) | | Descriptor: | (4-CARBAMIMIDOYLPHENYL)-METHYL-PHOSPHINIC ACID, CALCIUM ION, Trypsinogen | | Authors: | Cui, J, Marankan, F, Fu, W, Crich, D, Mesecar, A, Johnson, M.E. | | Deposit date: | 2004-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An oxyanion-hole selective serine protease inhibitor in complex with trypsin.
Bioorg.Med.Chem., 10, 2002
|
|
1U8V
 
 | | Crystal Structure of 4-Hydroxybutyryl-CoA Dehydratase from Clostridium aminobutyricum: Radical catalysis involving a [4Fe-4S] cluster and flavin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Gamma-aminobutyrate metabolism dehydratase/isomerase, IRON/SULFUR CLUSTER | | Authors: | Martins, B.M, Dobbek, H, Cinkaya, I, Buckel, W, Messerschmidt, A. | | Deposit date: | 2004-08-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of 4-hydroxybutyryl-CoA dehydratase: radical catalysis involving a [4Fe-4S] cluster and flavin.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2N7K
 
 | | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8 | | Descriptor: | NEDD8, Protein KHNYN | | Authors: | Santonico, E, Nepravishta, R, Mattioni, A, Valentini, E, Mandaliti, W, Procopio, R, Iannuccelli, M, Castagnoli, L, Polo, S, Paci, M, Cesareni, G. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8
To be Published
|
|
5IPI
 
 | | Structure of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
2NB7
 
 | |
7YC7
 
 | | Dark, fully reduced structure of the MmCPDII-DNA complex as produced at SwissFEL | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD7
 
 | | TR-SFX MmCPDII-DNA complex: 1 ns snapshot. Includes 1 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCM
 
 | | TR-SFX MmCPDII-DNA complex: 100 ps snapshot. Includes 100ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
