6TJL
 
 | |
6TJX
 
 | | Cryo-EM structure of TypeII tau filaments extracted from the brains of individuals with Corticobasal degeneration | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Murzin, A.G, Falcon, B, Shi, Y, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Novel tau filament fold in corticobasal degeneration.
Nature, 580, 2020
|
|
6TN6
 
 | | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | da Silva, V.M, Squina, F.M, Sperenca, M, Martin, L, Muniz, J.R.C, Garcia, W, Nicolet, Y. | | Deposit date: | 2019-12-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution structure of a modular hyperthermostable endo-beta-1,4-mannanase from Thermotoga petrophila: The ancillary immunoglobulin-like module is a thermostabilizing domain.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
6TSI
 
 | |
6L5A
 
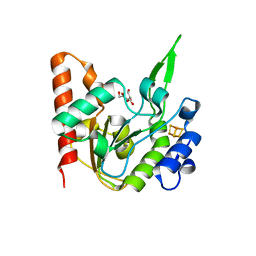 | | The structure of the UdgX mutant H109E at a pre-excision state | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Xie, W, Tu, J, Zeng, H. | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.80007327 Å) | | Cite: | Structural insights into an MsmUdgX mutant capable of both crosslinking and uracil excision capability.
DNA Repair (Amst), 97, 2021
|
|
6TRU
 
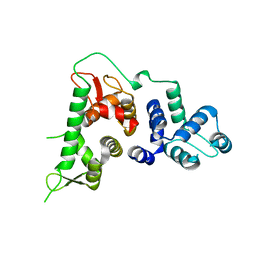 | |
6TTM
 
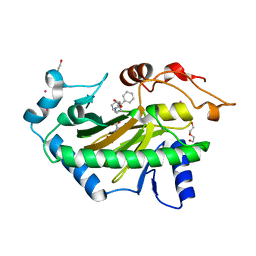 | | Hyoscyamine 6-hydroxylase in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyoscyamine 6 beta-hydroxylase, ... | | Authors: | Kluza, A, Kurpiewska, K, Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
6TU9
 
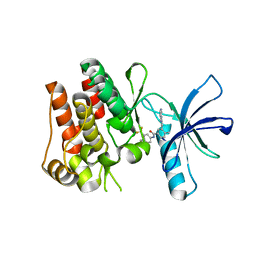 | | The ROR1 Pseudokinase Domain Bound To Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Niininen, W, Ungureanu, D, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
1A0S
 
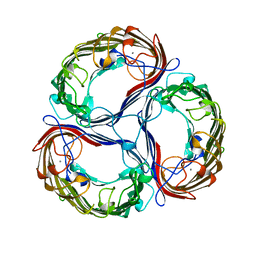 | | SUCROSE-SPECIFIC PORIN | | Descriptor: | CALCIUM ION, SUCROSE-SPECIFIC PORIN | | Authors: | Diederichs, K, Welte, W. | | Deposit date: | 1997-12-07 | | Release date: | 1998-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose.
Nat.Struct.Biol., 5, 1998
|
|
6TV2
 
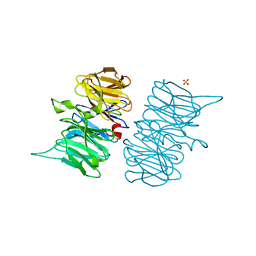 | | Heme d1 biosynthesis associated Protein NirF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Protein NirF, ... | | Authors: | Kluenemann, T, Layer, G, Blankenfeldt, W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Crystal structure of NirF: insights into its role in heme d 1 biosynthesis.
Febs J., 288, 2021
|
|
6TYD
 
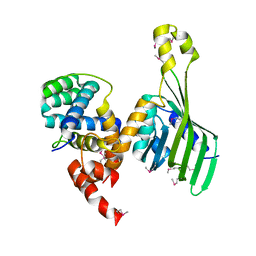 | | Structure of human LDB1 in complex with SSBP2 | | Descriptor: | LIM domain-binding protein 1, Single-stranded DNA-binding protein 2 | | Authors: | Wang, H, Wang, Z, Xu, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of human LDB1 in complex with SSBP2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1A0T
 
 | | SUCROSE-SPECIFIC PORIN, WITH BOUND SUCROSE MOLECULES | | Descriptor: | CALCIUM ION, SUCROSE-SPECIFIC PORIN, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Diederichs, K, Welte, W. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose.
Nat.Struct.Biol., 5, 1998
|
|
6U4X
 
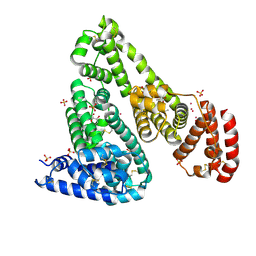 | | Crystal structure of Equine Serum Albumin complex with ibuprofen | | Descriptor: | IBUPROFEN, SULFATE ION, Serum albumin, ... | | Authors: | Czub, M.P, Handing, K.B, Venkataramany, B.S, Steen, E.H, Shabalin, I.G, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-26 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 63, 2020
|
|
6U7Z
 
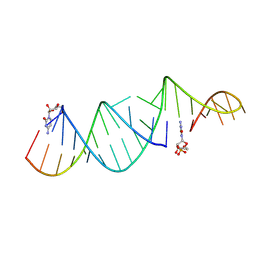 | | RNA hairpin structure containing one TNA nucleotide as primer | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*GP*CP*CP*GP*AP*AP*AP*GP*(TG))-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6UC4
 
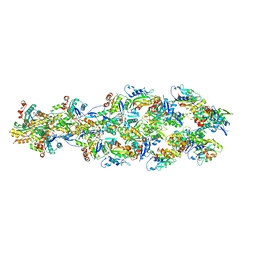 | | Barbed end side of a cofilactin cluster | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TV9
 
 | |
6LOI
 
 | |
1A5I
 
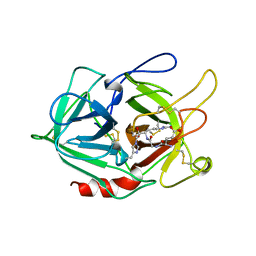 | |
1AQJ
 
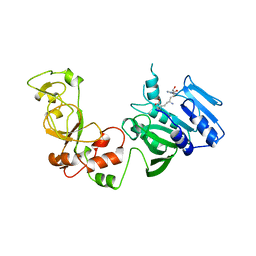 | | STRUCTURE OF ADENINE-N6-DNA-METHYLTRANSFERASE TAQI | | Descriptor: | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI, SINEFUNGIN | | Authors: | Schluckebier, G, Saenger, W. | | Deposit date: | 1996-07-25 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI.
J.Mol.Biol., 265, 1997
|
|
7RTC
 
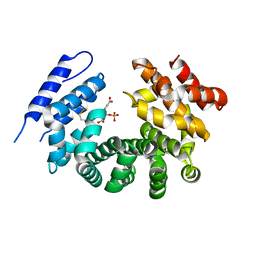 | |
5HCN
 
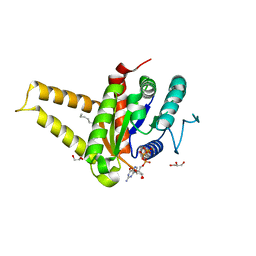 | | GPN-loop GTPase Npa3 in complex with GMPPCP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, LAURIC ACID, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
5HCI
 
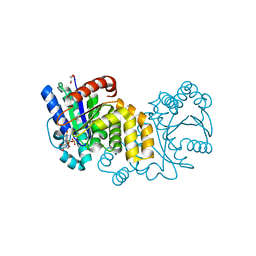 | | GPN-loop GTPase Npa3 in complex with GDP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
7RFR
 
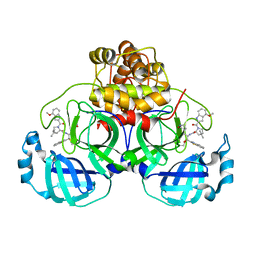 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFU
 
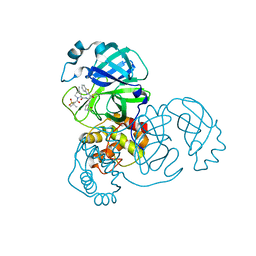 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFW
 
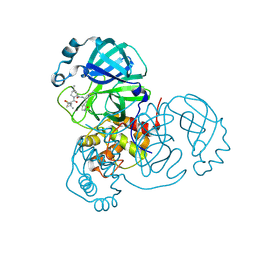 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
