7GKP
 
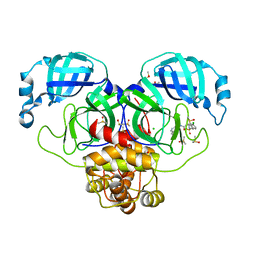 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-4223bc15-3 (Mpro-P1007) | | 分子名称: | (4S)-6-chloro-2-(cyclopropanesulfonyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.794 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GJA
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-de59a476-4 (Mpro-P0187) | | 分子名称: | (2R)-2-(3,4-dichlorophenyl)-N-(isoquinolin-4-yl)-2-(2-methoxyethoxy)acetamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.097 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6BHI
 
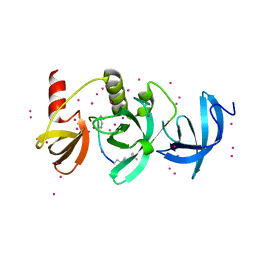 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
1FMK
 
 | |
1LCS
 
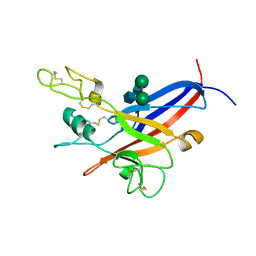 | | RECEPTOR-BINDING DOMAIN FROM SUBGROUP B FELINE LEUKEMIA VIRUS | | 分子名称: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Barnett, A.L, Wensel, D.L, Li, W, Fass, D, Cunningham, J.M. | | 登録日 | 2002-04-06 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and Mechanism of a Coreceptor for Infection by a pathogenic feline retrovirus
J.Virol., 77, 2003
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | 分子名称: | Anoctamin-1, CALCIUM ION | | 著者 | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | 登録日 | 2017-10-28 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BHD
 
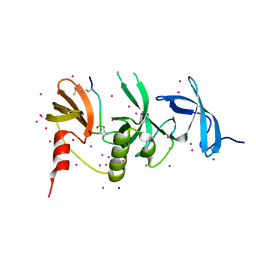 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SODIUM ION, ... | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
1FAV
 
 | | THE STRUCTURE OF AN HIV-1 SPECIFIC CELL ENTRY INHIBITOR IN COMPLEX WITH THE HIV-1 GP41 TRIMERIC CORE | | 分子名称: | HIV-1 ENVELOPE PROTEIN CHIMERA, PROTEIN (TRANSMEMBRANE GLYCOPROTEIN) | | 著者 | Zhou, G, Ferrer, M, Chopra, R, Strassmaier, T, Weissenhorn, W, Skehel, J.J, Oprian, D, Schreiber, S.L, Harrison, S.C, Wiley, D.C. | | 登録日 | 2000-07-13 | | 公開日 | 2000-08-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of an HIV-1 specific cell entry inhibitor in complex with the HIV-1 gp41 trimeric core.
Bioorg.Med.Chem., 8, 2000
|
|
1KJX
 
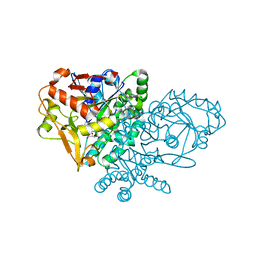 | | IMP Complex of E. Coli Adenylosuccinate Synthetase | | 分子名称: | Adenylosuccinate Synthetase, INOSINIC ACID | | 著者 | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | 登録日 | 2001-12-05 | | 公開日 | 2002-03-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
6C8E
 
 | | RNA-imidazolium-bridged intermediate complex, 4h soaking | | 分子名称: | 2-amino-1-[(R)-{[(2R,3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]-3-[(S)-{[(2R,3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]-1H-imidazol-3-ium, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G)-3') | | 著者 | Zhang, W, Szostak, J.W. | | 登録日 | 2018-01-24 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic observation of nonenzymatic RNA primer extension.
Elife, 7, 2018
|
|
1KLJ
 
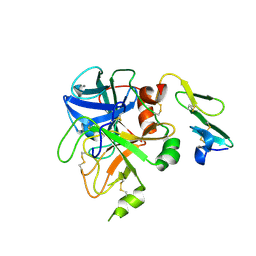 | | Crystal structure of uninhibited factor VIIa | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | 著者 | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | 登録日 | 2001-12-12 | | 公開日 | 2002-10-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
1KLY
 
 | |
1KM4
 
 | |
1KMC
 
 | |
6C2V
 
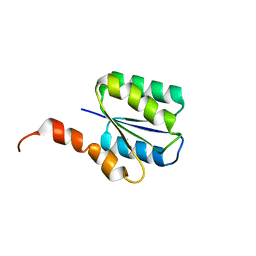 | | Solution structure of a phosphate-loop protein | | 分子名称: | phosphate-loop protein | | 著者 | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | 登録日 | 2018-01-09 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1KHF
 
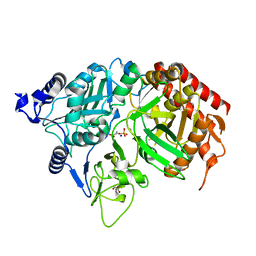 | | PEPCK complex with PEP | | 分子名称: | 1,2-ETHANEDIOL, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | 著者 | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | 登録日 | 2001-11-29 | | 公開日 | 2002-02-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1CFA
 
 | | SOLUTION STRUCTURE OF A SEMI-SYNTHETIC C5A RECEPTOR ANTAGONIST AT PH 5.2, 303K, NMR, 20 STRUCTURES | | 分子名称: | COMPLEMENT 5A SEMI-SYNTHETIC ANTAGONIST, SYNTHETIC N-TERMINAL TAIL | | 著者 | Zhang, X, Boyar, W, Galakatos, N, Gonnella, N.C. | | 登録日 | 1996-09-21 | | 公開日 | 1997-09-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a unique C5a semi-synthetic antagonist: implications in receptor binding.
Protein Sci., 6, 1997
|
|
1KKD
 
 | | Solution structure of the calmodulin binding domain (CaMBD) of small conductance Ca2+-activated potassium channels (SK2) | | 分子名称: | Small conductance calcium-activated potassium channel protein 2 | | 著者 | Wissmann, R, Bildl, W, Neumann, H, Rivard, A.F, Kloecker, N, Weitz, D, Schulte, U, Adelman, J.P, Bentrop, D, Fakler, B. | | 登録日 | 2001-12-07 | | 公開日 | 2001-12-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A helical region in the C terminus of small-conductance Ca2+-activated K+ channels controls assembly with apo-calmodulin.
J.Biol.Chem., 277, 2002
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | 分子名称: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-06 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
7GB8
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-14 (Mpro-x10247) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-[3-(trifluoromethyl)phenyl]acetamide | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.957 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6BLP
 
 | |
1KFM
 
 | |
1DF4
 
 | |
1KWP
 
 | | Crystal Structure of MAPKAP2 | | 分子名称: | MAP Kinase Activated Protein Kinase 2, MERCURY (II) ION | | 著者 | Meng, W, Swenson, L.L, Fitzgibbon, M.J, Hayakawa, K, ter Haar, E, Behrens, A.E, Fulghum, J.R, Lippke, J.A. | | 登録日 | 2002-01-30 | | 公開日 | 2002-09-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Mitogen-activated Protein Kinase-activated Protein (MAPKAP) Kinase 2 Suggests a Bifunctional Switch That
Couples Kinase Activation with Nuclear Export
J.Biol.Chem., 277, 2002
|
|
1DFY
 
 | | NMR STRUCTURE OF CONTRYPHAN-SM CYCLIC PEPTIDE (MAJOR FORM-CIS) | | 分子名称: | CONTRYPHAN-SM | | 著者 | Pallaghy, P.K, He, W, Jimenez, E.C, Olivera, B.M, Norton, R.S. | | 登録日 | 1999-11-22 | | 公開日 | 2002-05-01 | | 最終更新日 | 2020-06-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the contryphan family of cyclic peptides. Role of electrostatic interactions in cis-trans isomerism.
Biochemistry, 39, 2000
|
|
