4RDJ
 
 | | Crystal structure of Norovirus Boxer P domain | | 分子名称: | Capsid | | 著者 | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | 登録日 | 2014-09-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
2O52
 
 | | Crystal structure of human RAB4B in complex with GDP | | 分子名称: | BETA-MERCAPTOETHANOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Zhu, H, Tempel, W, Wang, J, Shen, Y, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2006-12-05 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of human RAB4B in complex with GDP
To be Published
|
|
5IHY
 
 | |
3KLK
 
 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in triclinic apo- form | | 分子名称: | CALCIUM ION, GLYCEROL, Glucansucrase | | 著者 | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | 登録日 | 2009-11-08 | | 公開日 | 2010-11-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KMR
 
 | | Crystal structure of RARalpha ligand binding domain in complex with an agonist ligand (Am580) and a coactivator fragment | | 分子名称: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | 著者 | Bourguet, W, Teyssier, C. | | 登録日 | 2009-11-11 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2NZV
 
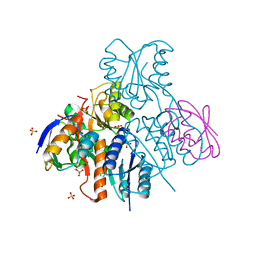 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | 著者 | Schumacher, M.A, Hillen, W, Brennan, R.G. | | 登録日 | 2006-11-25 | | 公開日 | 2007-05-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
4N70
 
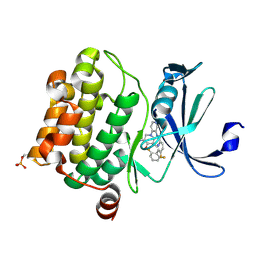 | | Pim1 Complexed with a pyridylcarboxamide | | 分子名称: | N-{4-[(3R,4R,5S)-3-amino-4-hydroxy-5-methylpiperidin-1-yl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | 著者 | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | 登録日 | 2013-10-14 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
6HH0
 
 | |
6BLP
 
 | |
4QV7
 
 | | yCP beta5-A50V mutant | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-14 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
2OD1
 
 | | Solution structure of the MYND domain from human AML1-ETO | | 分子名称: | Protein CBFA2T1, ZINC ION | | 著者 | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | 登録日 | 2006-12-21 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
4QW0
 
 | | yCP beta5-A49T-A50V-double mutant in complex with bortezomib | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-16 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4N9K
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with cephaloridine | | 分子名称: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase | | 著者 | Pan, X, Wong, W, Zhao, Y. | | 登録日 | 2013-10-21 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
6HOG
 
 | | Structure of VPS34 LIR motif bound to GABARAP | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, ... | | 著者 | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | 登録日 | 2018-09-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
2NSW
 
 | |
4QZ5
 
 | | yCP beta5-A49T-mutant in complex with ONX 0914 | | 分子名称: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
2O0Y
 
 | | Crystal structure of putative transcriptional regulator RHA1_ro06953 (IclR-family) from Rhodococcus sp. | | 分子名称: | Transcriptional regulator | | 著者 | Chruszcz, M, Wang, S, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-11-28 | | 公開日 | 2006-12-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Putative Transcriptional Regul 2 Rha1_Ro06953(Iclr-Family) from Rhodococcus Sp.
To be Published
|
|
2O20
 
 | | Crystal structure of transcription regulator CcpA of Lactococcus lactis | | 分子名称: | CHLORIDE ION, Catabolite control protein A, SULFATE ION | | 著者 | Loll, B, Kowalczyk, M, Alings, C, Chieduch, A, Bardowski, J, Saenger, W, Biesiadka, J. | | 登録日 | 2006-11-29 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the transcription regulator CcpA from Lactococcus lactis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6BPI
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates | | 分子名称: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, MLY-SER-THR-E2G, ... | | 著者 | MADER, P, Mendoza-Sanchez, R, DONG, A, DOBROVETSKY, E, IQBAL, A, CORLESS, V, TEMPEL, W, LIEW, S.K, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D.B, SCHAPIRA, M, VEDADI, M, BROWN, P.J, Santhakumar, V, FRYE, S, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-11-23 | | 公開日 | 2017-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates
to be published
|
|
2O3J
 
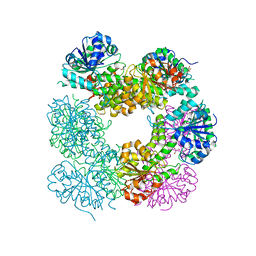 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | 分子名称: | GLYCEROL, UDP-glucose 6-dehydrogenase | | 著者 | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-12-01 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
6BSH
 
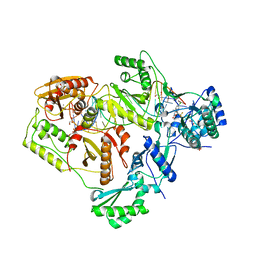 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in the RNA hydrolysis mode | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*AP*CP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | 著者 | Tian, L, Kim, M, Yang, W. | | 登録日 | 2017-12-03 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.649 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6IMC
 
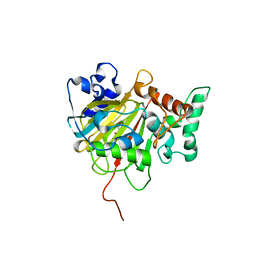 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | 分子名称: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | 著者 | Zhang, M, Yang, S, Zhao, W, Li, H. | | 登録日 | 2018-10-22 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
3KEM
 
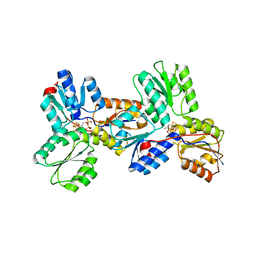 | | Crystal structure of IspH:IPP complex | | 分子名称: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | 著者 | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | 登録日 | 2009-10-26 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2OBL
 
 | | Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria | | 分子名称: | ACETATE ION, CALCIUM ION, EscN, ... | | 著者 | Zarivach, R, Vuckovic, M, Deng, W, Finlay, B.B, Strynadka, N.C.J. | | 登録日 | 2006-12-19 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural analysis of a prototypical ATPase from the type III secretion system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6INC
 
 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | 分子名称: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | 著者 | Wu, W, Zhang, Q, Bartlam, M. | | 登録日 | 2018-10-24 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.604 Å) | | 主引用文献 | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
