5Y9O
 
 | |
6BL1
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Direct Covalent Modification of His315 with Cmpd13 | | 分子名称: | (6aS,7S,9S,10aS)-7-methyl-8-oxo-10a-phenyl-2-(phenylamino)-5,6,6a,7,8,9,10,10a-octahydrobenzo[h]quinazoline-9-carbonitrile, CALCIUM ION, ISOCITRIC ACID, ... | | 著者 | Jakob, C.G, Qiu, W. | | 登録日 | 2017-11-09 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
1U1X
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | 分子名称: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | 著者 | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | 登録日 | 2004-07-16 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U2K
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (I41) | | 分子名称: | Peroxidase/catalase HPI | | 著者 | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | 登録日 | 2004-07-19 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6BM4
 
 | | Pol II elongation complex with an abasic lesion at i-1 position,soaking UMPNPP | | 分子名称: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DNA (5'-D(P*CP*AP*(3DR)P*CP*TP*CP*TP*TP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Wang, W, Wang, D. | | 登録日 | 2017-11-13 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.951 Å) | | 主引用文献 | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BKZ
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Non-equivalent Allosteric Inhibition with Cmpd3 | | 分子名称: | (7R)-1-[(4-fluorophenyl)methyl]-N-{3-[(1R)-1-hydroxyethyl]phenyl}-7-methyl-5-(1H-pyrrole-2-carbonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridine-3-carboxamide, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Jakob, C.G, Qiu, W. | | 登録日 | 2017-11-09 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
1YSM
 
 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | 分子名称: | Calcyclin-binding protein | | 著者 | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | 登録日 | 2005-02-08 | | 公開日 | 2005-07-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
6MKJ
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the closed conformation | | 分子名称: | penicillin binding protein 5 (PBP5) | | 著者 | Moon, T.M, Soares, A, D'Andrea, E.D, Jaconcic, J, Peti, W, Page, R. | | 登録日 | 2018-09-25 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.864 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
1G27
 
 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | 分子名称: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | 著者 | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | 登録日 | 2000-10-17 | | 公開日 | 2001-10-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G2P
 
 | | CRYSTAL STRUCTURE OF ADENINE PHOSPHORIBOSYLTRANSFERASE | | 分子名称: | ADENINE PHOSPHORIBOSYLTRANSFERASE 1, SULFATE ION | | 著者 | Shi, W, Tanaka, K.S.E, Almo, S.C, Schramm, V.L. | | 登録日 | 2000-10-20 | | 公開日 | 2001-12-05 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural analysis of adenine phosphoribosyltransferase from Saccharomyces cerevisiae.
Biochemistry, 40, 2001
|
|
1YN6
 
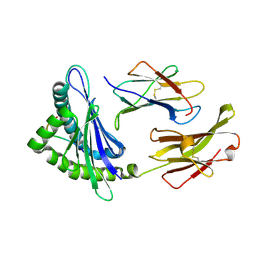 | | Crystal structure of a mouse MHC class I protein, H2-Db, in complex with a peptide from the influenza A acid polymerase | | 分子名称: | 10-mer peptide from RNA-directed RNA polymerase subunit P2, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Turner, S.J, Kedzierska, K, Komodromou, H, La Gruta, N.L, Dunstone, M.A, Webb, A.I, Webby, R, Walden, H, Xie, W, McCluskey, J, Purcell, A.W, Rossjohn, J, Doherty, P.C. | | 登録日 | 2005-01-23 | | 公開日 | 2005-06-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations
Nat.Immunol., 6, 2005
|
|
6BSG
 
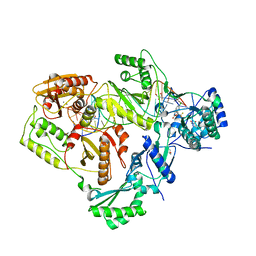 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in an RNA hydrolysis-off mode | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tian, L, Kim, M, Yang, W. | | 登録日 | 2017-12-03 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1GI2
 
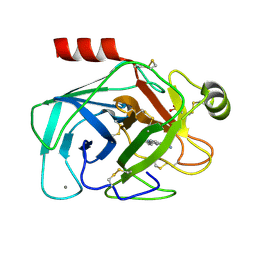 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-HYDROXY-PHENYL)-3H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1YOM
 
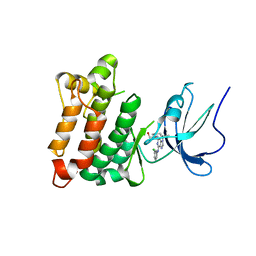 | | Crystal structure of Src kinase domain in complex with Purvalanol A | | 分子名称: | 2-({6-[(3-CHLOROPHENYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)-3-METHYLBUTAN-1-OL, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | 登録日 | 2005-01-27 | | 公開日 | 2006-01-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
1YQK
 
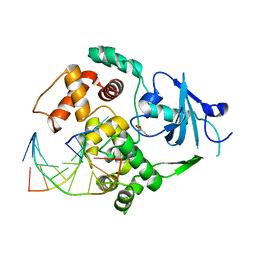 | | Human 8-oxoguanine glycosylase crosslinked with guanine containing DNA | | 分子名称: | 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*G)-3', 5'-D(P*CP*AP*GP*GP*TP*CP*TP*AP*C)-3', CALCIUM ION, ... | | 著者 | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | 登録日 | 2005-02-01 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
1YQR
 
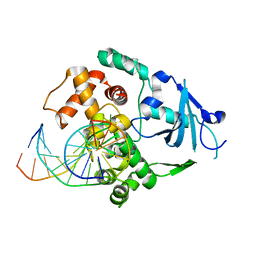 | | Catalytically inactive human 8-oxoguanine glycosylase crosslinked to oxoG containing DNA | | 分子名称: | 5'-D(P*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3', CALCIUM ION, ... | | 著者 | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | 登録日 | 2005-02-02 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
4FXX
 
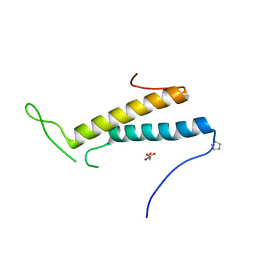 | | Structure of SF1 coiled-coil domain | | 分子名称: | IMIDAZOLE, MALONATE ION, Splicing factor 1 | | 著者 | Gupta, A, Bauer, W.J, Wang, W, Kielkopf, C.L. | | 登録日 | 2012-07-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4801 Å) | | 主引用文献 | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
6CER
 
 | | Human pyruvate dehydrogenase complex E1 component V138M mutation | | 分子名称: | MAGNESIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | 著者 | Whitley, M.J, Arjunan, P, Furey, W. | | 登録日 | 2018-02-12 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Pyruvate dehydrogenase complex deficiency is linked to regulatory loop disorder in the alpha V138M variant of human pyruvate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
1G8Y
 
 | | CRYSTAL STRUCTURE OF THE HEXAMERIC REPLICATIVE HELICASE REPA OF PLASMID RSF1010 | | 分子名称: | REGULATORY PROTEIN REPA | | 著者 | Niedenzu, T, Roeleke, D, Bains, G, Scherzinger, E, Saenger, W. | | 登録日 | 2000-11-21 | | 公開日 | 2001-02-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010.
J.Mol.Biol., 306, 2001
|
|
6MTG
 
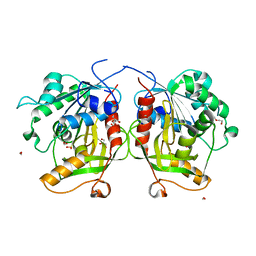 | | A Single Reactive Noncanonical Amino Acid is Able to Dramatically Stabilize Protein Structure | | 分子名称: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, GLYCEROL, ... | | 著者 | Li, J.C, Nasertorabi, F, Xuan, W, Han, G.W, Stevens, R.C, Schultz, P.G. | | 登録日 | 2018-10-19 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Single Reactive Noncanonical Amino Acid Is Able to Dramatically Stabilize Protein Structure.
Acs Chem.Biol., 14, 2019
|
|
1TOV
 
 | | Structural genomics of Caenorhabditis elegans: CAP-GLY domain of F53F4.3 | | 分子名称: | Hypothetical protein F53F4.3 in chromosome V, SULFATE ION | | 著者 | Li, S, Finley, J, Liu, Z.J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, Delucas, L.J, Richardson, D, Richardson, J, Wang, B.C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-06-15 | | 公開日 | 2004-07-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal Structure of the Cytoskeleton-Associated Protein Glycine-Rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
2MVG
 
 | |
1YOP
 
 | | The solution structure of Kti11p | | 分子名称: | Kti11p, ZINC ION | | 著者 | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | 登録日 | 2005-01-28 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
4GF6
 
 | | crystal structure of GFP with cuprum bound at the Incorporated metal Chelating Amino Acid PYZ151 | | 分子名称: | CALCIUM ION, COPPER (II) ION, green fluorescent protein | | 著者 | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | 登録日 | 2012-08-03 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1FYD
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE AMP, ONE PYROPHOSPHATE ION AND ONE MG2+ ION | | 分子名称: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | 著者 | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | 登録日 | 2000-09-28 | | 公開日 | 2001-06-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
