4C0C
 
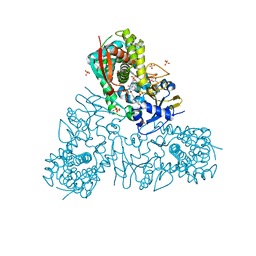 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(2,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | 4-[4-[2,4-bis(fluoranyl)phenyl]piperazin-1-yl]-2-fluoranyl-N-[(2R)-3-(1H-indol-3-yl)-1-oxidanylidene-1-(pyridin-4-ylamino)propan-2-yl]benzamide, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
3LEY
 
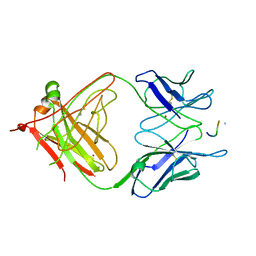 | | 2F5 Epitope scaffold elicited anti-HIV-1 monoclonal antibody 6a7 in complex with HIV-1 GP41 | | Descriptor: | 6a7 Antibody Heavy Chain, 6a7 Antibody Light Chain, Envelope glycoprotein gp41, ... | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Baker, D, Wyatt, R, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4GM1
 
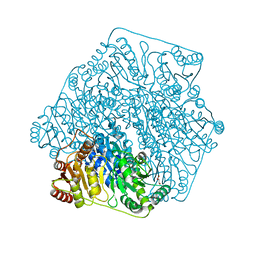 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403S | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
4GG1
 
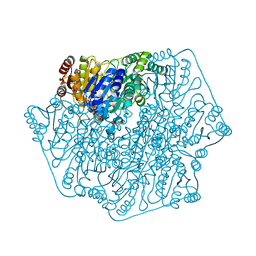 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403T | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-04 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
3LEX
 
 | | 2F5 Epitope scaffold elicited anti-HIV-1 monoclonal antibody 11F10 in complex with HIV-1 GP41 | | Descriptor: | 11f10 Antibody Heavy Chain, 11f10 Antibody Light Chain, Envelope glycoprotein gp41 | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Baker, D, Wyatt, R, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2CNU
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with aspartic acid | | Descriptor: | ASPARTIC ACID, PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-24 | | Release date: | 2006-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
2CNQ
 
 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with ADP, AICAR, succinate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
4CAT
 
 | | THREE-DIMENSIONAL STRUCTURE OF CATALASE FROM PENICILLIUM VITALE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Melik-Adamyan, W.R, Barynin, V.V, Vagin, A.A, Grebenko, A.I. | | Deposit date: | 1983-02-24 | | Release date: | 1983-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of catalase from Penicillium vitale at 2.0 A resolution.
J.Mol.Biol., 188, 1986
|
|
4BMM
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-2',3, 5'-trifluoro-(1,1'-biphenyl)-4-carboxamide | | Descriptor: | 4-[2,5-bis(fluoranyl)phenyl]-2-fluoranyl-N-[(2R)-3-(1H-indol-3-yl)-1-oxidanylidene-1-(pyridin-4-ylamino)propan-2-yl]benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Choi, J.Y, Calvet, C.M, Gunatilleke, S.S, Roush, W.R, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2013-05-09 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
4FBM
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4FBL
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4GRJ
 
 | |
2C0W
 
 | | Molecular Structure of fd Filamentous Bacteriophage Refined with Respect to X-ray Fibre Diffraction | | Descriptor: | COAT PROTEIN B | | Authors: | Marvin, D.A, Welsh, L.C, Symmons, M.F, Scott, W.R.P, Straus, S.K. | | Deposit date: | 2005-09-08 | | Release date: | 2005-12-14 | | Last modified: | 2024-02-14 | | Method: | FIBER DIFFRACTION (3.2 Å) | | Cite: | Molecular Structure of Fd (F1, M13) Filamentous Bacteriophage Refined with Respect to X-Ray Fibre Diffraction and Solid-State NMR Data Supports Specific Models of Phage Assembly at the Bacterial Membrane.
J.Mol.Biol., 355, 2006
|
|
3M9K
 
 | | Crystal structure of human thioredoxin C69/73S double-mutant, oxidized form | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, SULFATE ION, Thioredoxin | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2010-03-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human thioredoxin revealing an unraveled helix and exposed S-nitrosation site.
Protein Sci., 19, 2010
|
|
4A69
 
 | | Structure of HDAC3 bound to corepressor and inositol tetraphosphate | | Descriptor: | ACETATE ION, D-MYO INOSITOL 1,4,5,6 TETRAKISPHOSPHATE, GLYCEROL, ... | | Authors: | Watson, P.J, Fairall, L, Santos, G.M, Schwabe, J.W.R. | | Deposit date: | 2011-11-01 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Hdac3 Bound to Co-Repressor and Inositol Tetraphosphate.
Nature, 481, 2012
|
|
3M10
 
 | | Substrate-free form of Arginine Kinase | | Descriptor: | Arginine kinase, SULFATE ION | | Authors: | Yousef, M.S, Clark, S.A, Pruett, P.K, Somasundaram, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Arginine kinase: joint crystallographic and NMR RDC analyses link substrate-associated motions to intrinsic flexibility.
J.Mol.Biol., 405, 2011
|
|
4COH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the sulfonamide derivative of the 4-aminopyridyl-based inhibitor | | Descriptor: | 2-fluoranyl-N-[(2R)-3-(1H-indol-3-yl)-1-oxidanylidene-1-(pyridin-4-ylamino)propan-2-yl]-4-(4-thiophen-2-ylsulfonylpiperazin-1-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Vieira, D.F, Choi, J.Y, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Expanding the Binding Envelope of Cyp51 Inhibitors Targeting Trypanosoma Cruzi with 4-Aminopyridyl-Based Sulfonamide Derivatives
Chembiochem, 15, 2014
|
|
4CP3
 
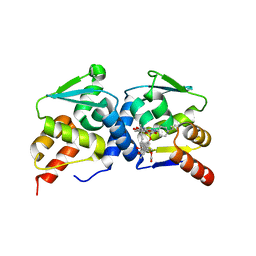 | | The structure of BCL6 BTB (POZ) domain in complex with the ansamycin antibiotic rifabutin. | | Descriptor: | B-CELL LYMPHOMA 6 PROTEIN, RIFABUTIN | | Authors: | Evans, S.E, Fairall, L, Goult, B.T, Jamieson, A.G, Ferrigno, P.K, Ford, R, Wagner, S.D, Schwabe, J.W.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Ansamycin Antibiotic, Rifamycin Sv, Inhibits Bcl6 Transcriptional Repression and Forms a Complex with the Bcl6-Btb/Poz Domain.
Plos One, 9, 2014
|
|
4GM0
 
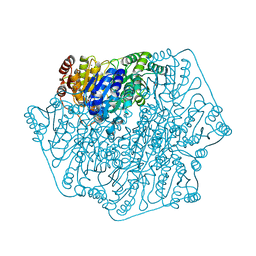 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403N | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
3NSY
 
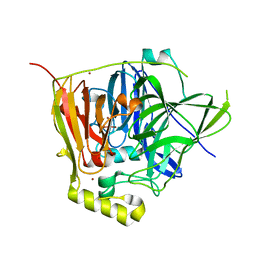 | | The multi-copper oxidase CueO with six Met to Ser mutations (M358S,M361S,M362S,M364S,M366S,M368S) | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
4GNW
 
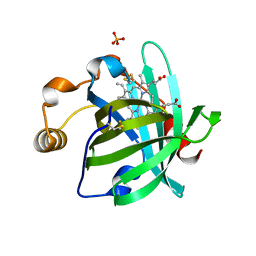 | |
4GPE
 
 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403M | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
3M9J
 
 | |
4C28
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(4-chlorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(4-chlorophenyl)piperazin-1-yl)-2-fluorobenzamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vieira, D.F, Calvet, C.M, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Binding Mode and Potency of N-Indolyl-Oxopyridinyl-4-Amino-Propanyl-Based Inhibitors Targeting Trypanosoma Cruzi Cyp51
J.Med.Chem., 57, 2014
|
|
2ALL
 
 | | Crystal structure of L122V/L132V mutant of nitrophorin 2 | | Descriptor: | Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-08-05 | | Release date: | 2006-07-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
