5SC1
 
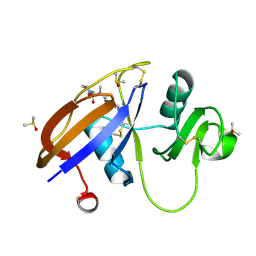 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z431807512 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.165 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SC2
 
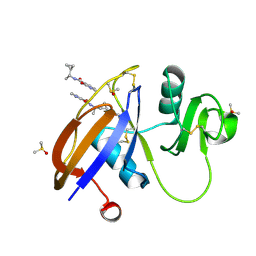 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z190780124 | | Descriptor: | (3S)-N-[(1R)-1-cyclopropylethyl]-2-oxo-2,3-dihydropyridine-3-carboxamide, 1,2-ETHANEDIOL, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SC3
 
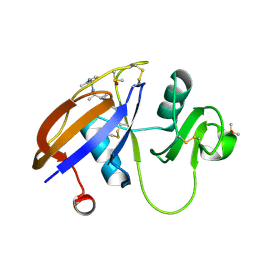 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z422471910 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SC4
 
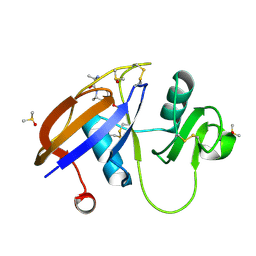 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z927412236 | | Descriptor: | (2R)-2-amino-1-[(2S)-2-methylpiperidin-1-yl]propan-1-one, 1,2-ETHANEDIOL, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.165 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SC5
 
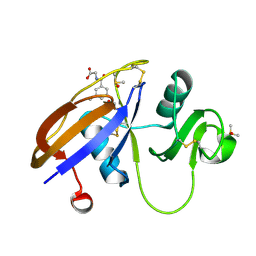 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z56827661 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SC6
 
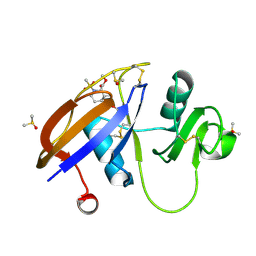 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with POB0019 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.327 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SC7
 
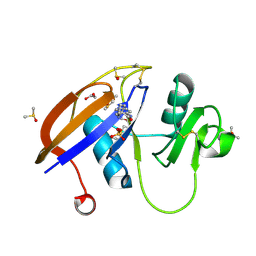 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with POB0120 | | Descriptor: | (2R)-1',4'-dihydro-2'H-spiro[pyrrolidine-2,3'-quinolin]-2'-one, 1,2-ETHANEDIOL, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.197 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5M9M
 
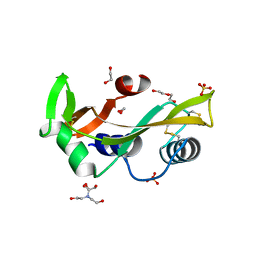 | | Human angiogenin PD variant Q77P | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Angiogenin, ... | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5NJL
 
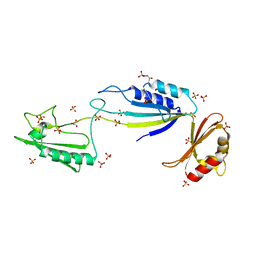 | | Cwp2 from Clostridium difficile | | Descriptor: | Cell surface protein (Putative S-layer protein), SULFATE ION | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-03-29 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cwp2 from Clostridium difficile exhibits an extended three domain fold and cell adhesion in vitro.
FEBS J., 284, 2017
|
|
5UI9
 
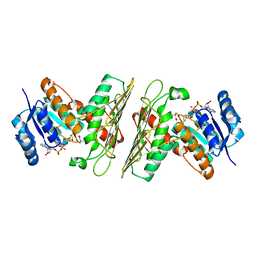 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, 2 -hydroxy-2-hydroxymethyl propanoic acid and Magnesium | | Descriptor: | (2S)-2,3-dihydroxy-2-methylpropanoic acid, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, 2 -hydroxy-2-hydroxymethyl propanoic acid and Magnesium
To be published
|
|
1QX2
 
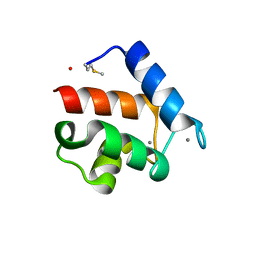 | | X-ray Structure of Calcium-loaded Calbindomodulin (A Calbindin D9k Re-engineered to Undergo a Conformational Opening) at 1.44 A Resolution | | Descriptor: | CALCIUM ION, Vitamin D-dependent calcium-binding protein, intestinal, ... | | Authors: | Bunick, C.G, Nelson, M.R, Mangahas, S, Mizoue, L.S, Bunick, G.J, Chazin, W.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-05-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Designing Sequence to Control Protein Function in an EF-Hand Protein
J.Am.Chem.Soc., 126, 2004
|
|
1R4Z
 
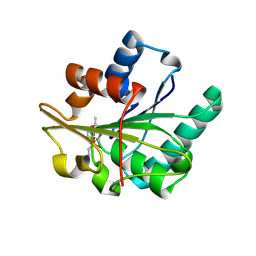 | | Bacillus subtilis lipase A with covalently bound Rc-IPG-phosphonate-inhibitor | | Descriptor: | Lipase, [(4R)-2,2-DIMETHYL-1,3-DIOXOLAN-4-YL]METHYL HYDROGEN HEX-5-ENYLPHOSPHONATE | | Authors: | Droege, M.J, Van Pouderoyen, G, Vrenken, T.E, Rueggeberg, C.J, Reetz, M.T, Dijkstra, B.W, Quax, W.J. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed Evolution of Bacillus subtilis Lipase A by Use of Enantiomeric Phosphonate Inhibitors: Crystal Structures and Phage Display Selection
Chembiochem, 7, 2005
|
|
1RD3
 
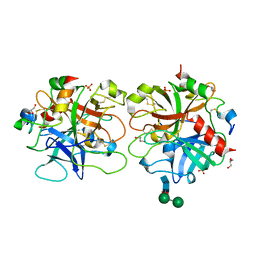 | | 2.5A Structure of Anticoagulant Thrombin Variant E217K | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Carter, W.J, Myles, T, Leung, L.L, Huntington, J.A. | | Deposit date: | 2003-11-05 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Anticoagulant Thrombin Variant E217K Provides Insights into Thrombin Allostery
J.Biol.Chem., 279, 2004
|
|
1RZ0
 
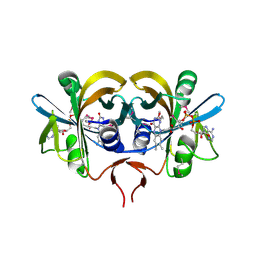 | | Flavin reductase PheA2 in native state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, phenol 2-hydroxylase component B | | Authors: | van den Heuvel, R.H, Westphal, A.H, Heck, A.J, Walsh, M.A, Rovida, S, van Berkel, W.J, Mattevi, A. | | Deposit date: | 2003-12-23 | | Release date: | 2004-04-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on Flavin Reductase PheA2 Reveal Binding of NAD in an Unusual Folded Conformation and Support Novel Mechanism of Action.
J.Biol.Chem., 279, 2004
|
|
1RO9
 
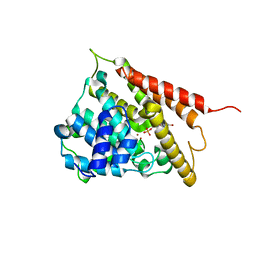 | | CRYSTAL STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHODIESTERASE 4B2B COMPLEXED WITH 8-Br-AMP | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4B | | Authors: | Xu, R.X, Rocque, W.J, Lambert, M.H, Vanderwall, D.E, Nolte, R.T. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram.
J.Mol.Biol., 337, 2004
|
|
1RRI
 
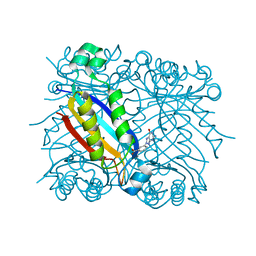 | | DHNA complex with 3-(5-amino-7-hydroxy-[1,2,3] triazolo [4,5-d]pyrimidin-2-yl)-benzoic acid | | Descriptor: | 3-(5-AMINO-7-HYDROXY-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-2-YL)-BENZOIC ACID, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
1SK6
 
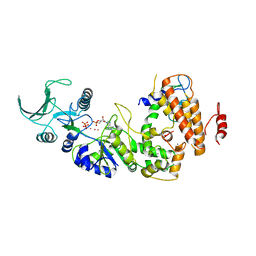 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin, 3',5' cyclic AMP (cAMP), and pyrophosphate | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Shen, Y, Zhukovskaya, N.L, Tang, W.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate.
J.Biol.Chem., 279, 2004
|
|
1SQA
 
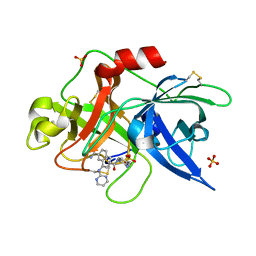 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-4-(PYRIMIDIN-2-YLAMINO)-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1SBO
 
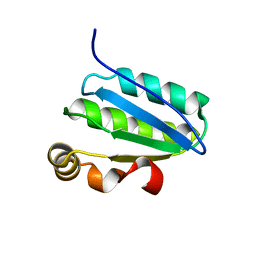 | | Solution Structure of putative anti sigma factor antagonist from Thermotoga maritima (TM1442) | | Descriptor: | Putative anti-sigma factor antagonist TM1442 | | Authors: | Etezady-Esfarjaini, T, Placzek, W.J, Herrmann, T, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the putative anti-sigma-factor antagonist TM1442 from Thermotoga maritima in the free and phosphorylated states.
Magn.Reson.Chem., 44 Spec No, 2006
|
|
1PX6
 
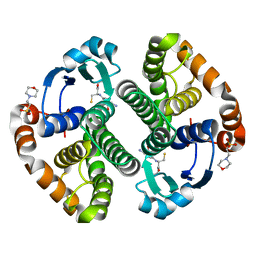 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to asparagine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase P | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1PSB
 
 | | Solution structure of calcium loaded S100B complexed to a peptide from N-Terminal regulatory domain of NDR kinase. | | Descriptor: | Ndr Ser/Thr kinase-like protein, S-100 protein, beta chain | | Authors: | Bhattacharya, S, Large, E, Heizmann, C.W, Hemmings, B, Chazin, W.J. | | Deposit date: | 2003-06-21 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ca(2+)/S100B/NDR Kinase Peptide Complex: Insights into S100 Target Specificity and Activation of the Kinase.
Biochemistry, 42, 2003
|
|
1SMT
 
 | | SMTB REPRESSOR FROM SYNECHOCOCCUS PCC7942 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR SMTB | | Authors: | Cook, W.J, Hall, L.M. | | Deposit date: | 1997-09-16 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins.
J.Mol.Biol., 275, 1998
|
|
1SQT
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 7-METHOXY-8-[1-(METHYLSULFONYL)-1H-PYRAZOL-4-YL]NAPHTHALENE-2-CARBOXIMIDAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhang, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1RZ1
 
 | | Reduced flavin reductase PheA2 in complex with NAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, phenol 2-hydroxylase component B | | Authors: | Van Den Heuvel, R.H, Westphal, A.H, Heck, A.J, Walsh, M.A, Rovida, S, Van Berkel, W.J, Mattevi, A. | | Deposit date: | 2003-12-23 | | Release date: | 2004-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies on Flavin Reductase PheA2 Reveal Binding of NAD in an Unusual Folded Conformation and Support Novel Mechanism of Action.
J.Biol.Chem., 279, 2004
|
|
1R50
 
 | | Bacillus subtilis lipase A with covalently bound Sc-IPG-phosphonate-inhibitor | | Descriptor: | Lipase, [(4S)-2,2-DIMETHYL-1,3-DIOXOLAN-4-YL]METHYL HYDROGEN HEX-5-ENYLPHOSPHONATE | | Authors: | Droege, M.J, Van Pouderoyen, G, Vrenken, T.E, Rueggeberg, C.J, Reetz, M.T, Dijkstra, B.W, Quax, W.J. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Directed Evolution of Bacillus subtilis Lipase A by Use of Enantiomeric Phosphonate Inhibitors: Crystal Structures and Phage Display Selection
Chembiochem, 7, 2005
|
|
