6B70
 
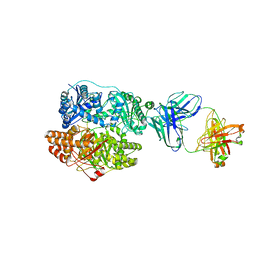 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin | | Descriptor: | FAB H11-E heavy chain, FAB H11-E light chain, Insulin, ... | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B2E
 
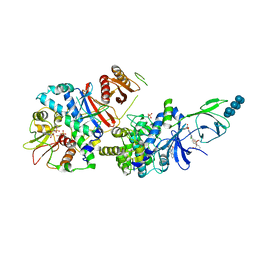 | | Structure of full length human AMPK (a2b2g1) in complex with a small molecule activator SC4. | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
6BF8
 
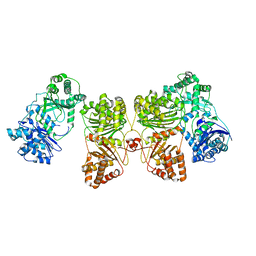 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B1U
 
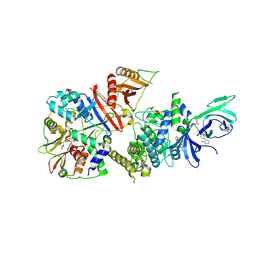 | | Structure of full-length human AMPK (a2b1g1) in complex with a small molecule activator SC4 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
6BFC
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-27 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B3Q
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | Descriptor: | Insulin, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-22 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Y
 
 | | Cryo-EM structure of human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF6
 
 | | Cryo-EM structure of human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Z
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | Descriptor: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-10 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
1F8X
 
 | |
6C6D
 
 | |
6DU0
 
 | |
1DLP
 
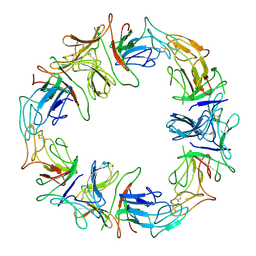 | | STRUCTURAL CHARACTERIZATION OF THE NATIVE FETUIN-BINDING PROTEIN SCILLA CAMPANULATA AGGLUTININ (SCAFET): A NOVEL TWO-DOMAIN LECTIN | | Descriptor: | LECTIN SCAFET PRECURSOR | | Authors: | Wright, L.M, Reynolds, C.D, Rizkallah, P.J, Allen, A.K, VanDamme, E.J.M, Donovan, M.J, Peumans, W.J. | | Deposit date: | 1999-12-11 | | Release date: | 2000-02-10 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of the native fetuin-binding protein Scilla campanulata agglutinin: a novel two-domain lectin.
FEBS Lett., 468, 2000
|
|
1HAF
 
 | |
1HAE
 
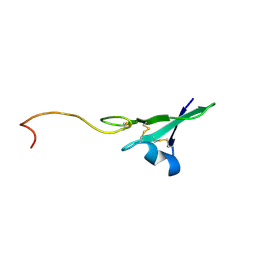 | |
1HOM
 
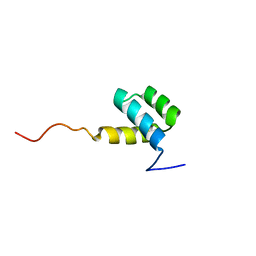 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Qian, Y.-Q, Billeter, M, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
5DUQ
 
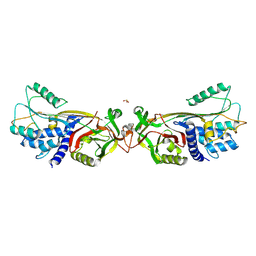 | | Active human c1-inhibitor in complex with dextran sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasma protease C1 inhibitor, SULFITE ION, ... | | Authors: | Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A, Pannu, N.S. | | Deposit date: | 2015-09-20 | | Release date: | 2016-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
5DQO
 
 | |
5EMW
 
 | | Crystal structure of the palmitoylated human TEAD3 transcription factor | | Descriptor: | CALCIUM ION, Transcriptional enhancer factor TEF-5 | | Authors: | Noland, C.L, Gierke, S, Schnier, P.D, Murray, J, Sandoval, W.N, Sagolla, M, Dey, A, Hannoush, R.N, Fairbrother, W.J, Cunningham, C.N. | | Deposit date: | 2015-11-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling.
Structure, 24, 2016
|
|
5FWI
 
 | |
5GSV
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GR7
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5EMV
 
 | | Crystal structure of the palmitoylated human TEAD2 transcription factor | | Descriptor: | Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Gierke, S, Schnier, P.D, Murray, J, Sandoval, W.N, Sagolla, M, Dey, A, Hannoush, R.N, Fairbrother, W.J, Cunningham, C.N. | | Deposit date: | 2015-11-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling.
Structure, 24, 2016
|
|
