6D03
 
 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; one molecule of parasite ligand. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(2-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6S9U
 
 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Ilumatobacter coccineus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
6MTQ
 
 | | Crystal structure of VRC42.N1 Fab in complex with T117-F MPER scaffold | | Descriptor: | Antibody VRC42.N1 Fab heavy chain, Antibody VRC42.N1 Fab light chain, VRC42 epitope T117-F scaffold | | Authors: | Kwon, Y.D, Law, W.H, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
4RM4
 
 | | The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhouw, W.H, Zhang, A.L, Zhang, T, Hall, E.A, Hutchinson, S, Cryle, M.J, Wong, L.-L, Bell, S.G. | | Deposit date: | 2014-10-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis.
Mol Biosyst, 11, 2015
|
|
8T56
 
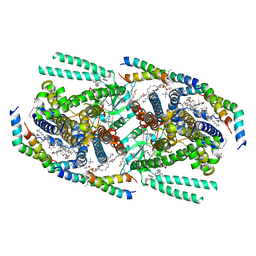 | | Structure of mechanically activated ion channel OSCA1.2 in peptidiscs | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Burendei, B, Jojoa-Cruz, S, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of mechanically activated ion channel OSCA2.3 reveals mobile elements in the transmembrane domain.
Structure, 32, 2024
|
|
8T57
 
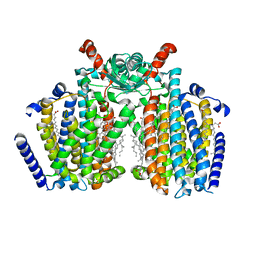 | | Structure of mechanically activated ion channel OSCA2.3 in peptidiscs | | Descriptor: | CHOLESTEROL, CSC1-like protein HYP1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Burendei, B, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of mechanically activated ion channel OSCA2.3 reveals mobile elements in the transmembrane domain.
Structure, 32, 2024
|
|
6D05
 
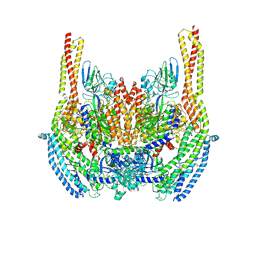 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6D04
 
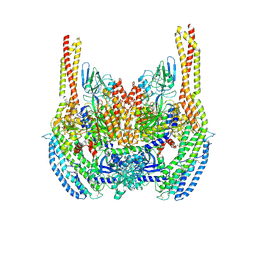 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
8SEF
 
 | | Crystal structure of Cy137C02, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine | | Descriptor: | Cy137C02 Fab heavy chain, Cy137C02 Fab light chain, GLYCEROL, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-09 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of Cy137C02, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine
To Be Published
|
|
8SGN
 
 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine
To Be Published
|
|
6BPA
 
 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 3E9 | | Descriptor: | BROMIDE ION, Monoclonal antibody 3E9 Fab heavy chain, Monoclonal antibody 3E9 Fab light chain, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
8SGA
 
 | | Crystal structure of 770E11, a monoclonal antibody isolated from a human Epstein-Barr virus seropositive donor | | Descriptor: | 770E11 Fab heavy chain, 770E11 Fab light chain, GLYCEROL, ... | | Authors: | Chen, W.H, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 770E11, a monoclonal antibody isolated from a human Epstein-Barr virus seropositive donor
To Be Published
|
|
8SIC
 
 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy137C02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cy137C02 Fab heavy chain, Cy137C02 Fab light chain, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy137C02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine
To Be Published
|
|
8SGG
 
 | | Crystal structure of Cy137D09, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine | | Descriptor: | Cy137D09 Fab heavy chain, Cy137D09 Fab light chain, GLYCEROL | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Cy137D09, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine
To Be Published
|
|
6R1L
 
 | |
6BPC
 
 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 4F7 | | Descriptor: | Monoclonal antibody 4F7 Fab heavy chain, Monoclonal antibody 4F7 Fab light chain, Reticulocyte binding protein 2, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
1UB1
 
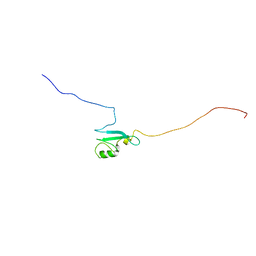 | | Solution structure of the matrix attachment region-binding domain of chicken MeCP2 | | Descriptor: | attachment region binding protein | | Authors: | Heitmann, B, Maurer, T, Weitzel, J.M, Stratling, W.H, Kalbitzer, H.R, Brunner, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the matrix attachment region-binding domain of chicken MeCP2
EUR.J.BIOCHEM., 270, 2003
|
|
4V4U
 
 | | The quasi-atomic model of Human Adenovirus type 5 capsid | | Descriptor: | HEXON PROTEIN, N-TERMINAL PEPTIDE OF FIBER PROTEIN, PENTON PROTEIN | | Authors: | Fabry, C.M.S, Rosa-Calatrava, M, Conway, J.F, Zubieta, C, Cusack, S, Ruigrok, R.W.H, Schoehn, G. | | Deposit date: | 2005-03-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | A Quasi-Atomic Model of Human Adenovirus Type 5 Capsid.
Embo J., 24, 2005
|
|
4UFT
 
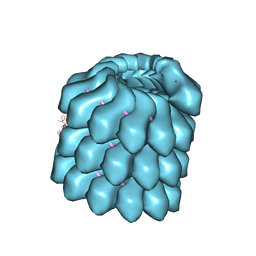 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
6BPE
 
 | | Plasmodium vivax reticulocyte binding protein 2b (PvRBP2b) bound to monoclonal antibody 6H1 | | Descriptor: | Monoclonal antibody 6H1 Fab heavy chain, Monoclonal antibody 6H1 Fab light chain, Reticulocyte binding protein 2, ... | | Authors: | Gruszczyk, J, Chan, L.J, Tham, W.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
4U5P
 
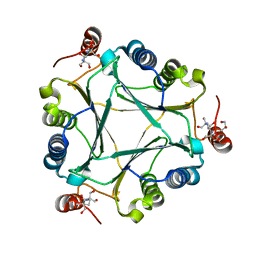 | | Crystal structure of native RhCC (YP_702633.1) from Rhodococcus jostii RHA1 at 1.78 Angstrom | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Poddar, H, Rozeboom, H.J, Thunnissen, A.M.W.H. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Functional and structural characterization of an unusual cofactor-independent oxygenase.
Biochemistry, 54, 2015
|
|
4U5R
 
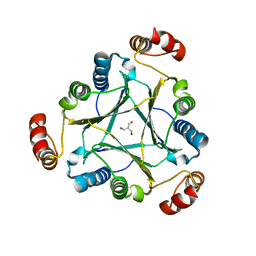 | |
6NF6
 
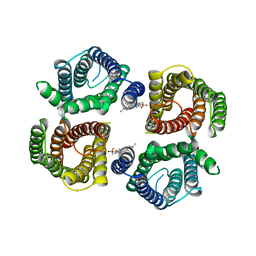 | | Structure of chicken Otop3 in nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Otopetrin3 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6RXD
 
 | |
6TB0
 
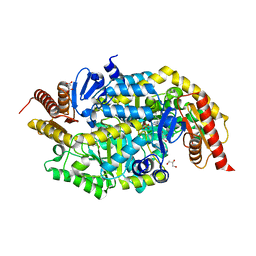 | | Crystal structure of thermostable omega transaminase 4-fold mutant from Pseudomonas jessenii | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
